Study on Genetic Factors of Otosclerosis: Review
- 1. Zhejiang University School of Medicine, Hangzhou, China
- 2. Department of Otolaryngology, The First Affiliated Hospital Zhejiang University School of Medicine, China
ABSTRACT
Otosclerosis is a primary metabolic bone disease, formed by abnormal bone remolding in the otic capsule, which results in progressive conductive hearing loss and may have sensorineural component when the cochlea is involved. The exact etiology of otosclerosis is unclear. As we know, osclerosis has obvious genetic tendency, 60% of them tends to cluster in families, which is deemed as autosomal dominant inheritance and variable expression. For decades, researchers have identified many disease-causing genes and diseaseassociated genes to illustrate the complex pathophysiology of otosclerosis. In this review, we summarized the studies on genetic factors of otosclerosis.
KEYWORDS
• Otosclerosis
• Hearing loss
• Genetics
• Genes
CITATION
Cai L, Xu Y, Liu Y, Chen Q (2022) Study on Genetic Factors of Otosclerosis: Review. Ann Otolaryngol Rhinol 9(3): 1289.
INTRODUCTION
Otosclerosis is a localized bone dysplasia disease in the otic capsule, a special bony structure which often undergoes less bone remodeling than the bones in other parts of human body [1,2]. Otosclerosis is formed by primary localized bone resorption of the bone labyrinth, and remolded by spongy bone hyperplasia with abundant blood vessels. Fixation of the stapes to the oval window is the mainly histological feature [1-5]. The lesion of otosclerosis is further involved in the bone labyrinth rather than invading the conduction and sensorineural structure, it is called histological otosclerosis. The histological otosclerosis patients will not suffer from clinical symptoms, such as hearing loss, tinnitus or vertigo, throughout their lives in which the disease only can be diagnosed by postmortem or by high-resolution computed tomographic scanning. The prevalence of clinical otosclerosis is about 0.3- 0.4% in white people, but there was no obvious difference in sex, while histological otosclerosis is almost 10 times more frequent than clinical otosclerosis. The frequency of otosclerosis also differs from race to race, and it is more common in white people than Asian and black people due to the early studies. The difference in frequency of otosclerosis between different races might result from the genetic and environmental differences. The clinical otosclerosis always manifests as progressive conductive hearing loss, while 10% of patients with sensorineural hearing loss or mixed hearing loss also exist. The bilateral cases account for about 80% of all cases, clinical symptoms always occur in their early thirties, and get worse during pregnancy [6-8].
The exact etiology of otosclerosis remains unclear up to now, while genetic, endocrine, immune and environmental factors are postulated to be mainly factors. Genetic factors is considered as most important, because 60% of clinical otosclerosis patients have a significant family history and Mendel’s law seems to be applied among a half of those patients with family history. The descendant of the otosclerosis patients are also at high risk of otosclerosis [9-11]. A recent family study has proved the exist of autosomal dominant inheritance and found a delay in onset age of the family cases. Besides, the other half cases without positive family history were reported in present of complex inheritance modes.
The paper made a review of the genetic factors of otosclerosis based on the study in recent years.
Disease-causing genes
Several linkage analyses have been performed over the years, and thousands of genetic markers were studied to determine the chromosome region responsible for the genes.. The hereditary pattern is incomplete penetrance, and there are eight genetic loci published currently, respectively on different chromosomes, as listed in the Table 1.
Table 1: Loci determined by linkage analysis.
|
Locus |
Position |
Author of the study |
|
OTSC1 |
15q25-q26 |
Tomek et al. |
|
OTSC2 |
7q34-36 |
Van Den Bogaert et al. |
|
OTSC3 |
6p21.3-22.3 |
Chen et al. |
|
OTSC4 |
16q21-23.2 |
Brownstein et al. |
|
OTSC5 |
3q22-24 |
Van Den Bogaert et al. |
|
OTSC6 |
unpublished |
|
|
OTSC7 |
6q13-16.1 |
Thys et al. |
|
OTSC8 |
9p13.1-q21.11 |
Bel Hadj Ali et al. |
|
OTSC9 |
unpublished |
|
|
OTSC10 |
1q41-44 |
Schrauwen et al. |
Early in 1998, Tomek et al. reported the first loci OTSC1, which was detected in the cases from the same family [12-19]. They found the elder cases were more serious in sensorineural hearing loss than the younger cases, while the conductive loss did not show any significant difference. Subsequent genetic linkage analysis via Short Tandem Repeat Polymorphisms (STRPs) resulted in the calculated maximum multipoint lod score of 3.4 [20]. Further linkage analysis pinpointed the region between the Far Centriole (FES) and the near centriole (D15S657), a 14.5cM (centimorgan) segment of chromosome 15’s long arm that may harbor the otosclerosis gene. Moreover, the FES–D15S657 interval is aggrecan, the major non-collagenous component in quantity of the cartilaginous extracellular matrix [21-28].
As for the other genes above, the OTSC2 region includes some known genes, such as TIF1a (transcription intermediary factor 1-alpha), PLOD3 (procollagen-lysine, 2-oxyglutarate, 5-dioxygenase 3) and TNFa (tumor necrosis factor–alpha) [29-35]. TIF1a is a growth inhibitor of retinoic acid, which disrupts the development and differentiation of the otic capsule. PLOD3 maps to the candidate region and takes part in collagen biosynthesis and metabolism. TNFa is a key mediator in the arthritis pathogenesis, causing the degradation of cartilage and the destruction of joints, and can enhance the activity of PLOD3 [36-42]. The OTSC3 region contains the HLA (human leukocyte antigen) region, which is consistent with the correlations between HLA-A/HLA-B antigens and otosclerosis reported before. The defined OTSC4 region involves several genes related to the immune system and bone homeostasis [43,44]. The OTSC5 region involves two supposed great candidate genes: PCOLCE2 (procollagen COOH-terminal proteinase enhancer protein 2) and CHST2 (carbohydrate sulfotransferase 2). However, the subsequent mutation analyses showed no disease-causing mutation of these two genes. The defined OTSC7 interval involves a credible candidate gene COL12A1 (collagen type II alpha 1) which belongs to the fibrilassociated collagens with discontinuous triple helices, and expresses in the cochlea, while the subsequent mutation analyses failed to reveal any disease-causing mutation. The OTSC8 region involves three candidate genes: TJP2 (tight junction protein 2), TRPM3 (transient receptor potential cation channel, subfamily M, member 3) and KLF9 (kruppel like factor 9). TJP2 belongs to the MAGUK (membrane-associated guanylate kinase) homologues family and takes part in epithelial and endothelial intracellular junctions. TRPM3 is a cation-selective channel, which takes an important part in cellular calcium signaling, homeostasis and osteoclast activity [45-52]. KLF9 is an effective activator of AP-2 (activating enhancer binding protein 2 alpha), a regulator of development in the mammalian craniofacial. The OTSC10 region involves 306 gene predictions including two candidate genes (Figure 1),
Figure 1: Gene predictions including candidate genes.
TGFB2 (transforming growth factor beta 2) and AGT (angiotensinogen), both of which play an important role in bone remodeling and were found associated to otosclerosis previously [53-58].
Though many loci involved, the disease-causing gene of each region remained undefined except for the T cell receptor beta, which was supposed to be the responsible gene at the OTSC2 region [59-61]. These loci have revealed the genetic heterogeneity of otosclerosis. However, about 40-50% of clinical cases, which seemed to be sporadic and did not follow the Mendel’s law, might result from the reduced penetrance, the environmental factors or other models of inheritance besides autosomal dominant.
Disease-associated candidate genes
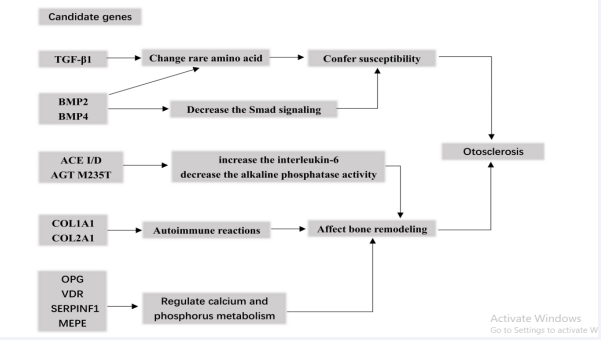
Collagen genes: COL2A1 gene was firstly analyzed, due to its relation to the globuli interossei and the hypothesis that autoimmune reactions to Type II collagen might exist in the progress of otosclerosis. Immune modulating factors as well as immune cells have been already proved to exest in otosclerosis foci. Though other lesions localized chondrodysplasia , there is no evidence to prove COL2A1 could be a cause of otosclerosis by subsequent studies [62].
The COL1A1 (collagen type I alpha 1) gene, which is involved in bone remodeling, was firstly found to be associated with otosclerosis in the the study by three different polymorphic markers within the gene by Mc Kenna et al. in 1998. Only a small percentage of mutations in COL1A1 were similar to the ones occurring in type I osteogenesis imperfecta, and further study showed an association between otosclerosis and the first intron Sp1 site of COL1A1 gene. A recent study of a Chinese otosclerosis family also showed the c.2209A > G (p.T737A) mutation of the SP1 gene in exon 6 may be the causative gene of otosclerosis, and responsible for the autosomal dominant inheritance. Increasing of COL1A1 homotrimer formation might have a relationship with the development of otosclerosis. In addition, COL1A1 and COL1A2 (collagen type I alpha 2) associate to form a collagen Type I triple helix in a 2:1 ratio in normal condition, and they demonstrated that some polymophisms can lead to increase the production of COL1A1 homomeric trimers by altering the binding of the transcription factors which regulates COL1A2 transcription. Several subsequent genetic studies also confirmed the evidence for polymorphisms of COL1A1, but some reported negative results. Schrauwen et al. found the evidence for the correlation between COL1A1 and otosclerosis by meta-analysis. However, they could not replicate any SNPs that were significant in earlier studies, the effect sizes of the variants reported before were probably an overestimate of true effect sizes.
Further, possible correlation of COL1A1 gene and otosclerosis will be describled by larger heterogeneous patient population.
TGF-β superfamily: The TGF-β1 (transforming growth factor-β1) belongs to the TGF-β superfamily, and is related to the bone metabolism of the otic capsule. TGFβ-1 has been confirmed to modify the phenotype of the otosclerotic cells, and associated with otosclerosis in independent population. In those studies, it was concluded that the protective multiple rare amino acid changing variants of TGFβ-1 may inhibit osteoclast differentiation and activation, and decrease the susceptibility to otosclerosis. Several other studies have also replicated and strengthened the evidence that the rs1800472 SNP of TGFβ-1 gene relate to otosclerosis. Later, a risk variant c.-509C > T and a risk haplotype G-T-T-G in the TGFβ-1 gene that may increase the risk to otosclerosis were revealed, and antoher TGFβ-1 mutation b-832G > A was identified to lead to increase the susceptibility to otosclerosis by altering the TGFβ-1 promoter activity. Although the mechanism of TGFβ-1 in the pathogenesis of otosclerosis is unclear, the theory that TGFβ-1 influences the globuli interossei within the otic capsule in the chondrogenesis process is supported by the data of the proteomic analysis.
BMP2 and BMP4 (bone morphogenetic protein 2/4) genes also belong to the TGF-beta superfamily, which were susceptible to otosclerosis in two large independent populations. The variant of SNP (rs3178250), located in the 3’ untranslated region of BMP2, may increase the susceptibility of otosclerosis. The variant of SNP (rs17563), located in BMP4, can also confers susceptibility by changing the amino acid. Furthemore, BMP2 and BMP4 might have the function of decreasing the Smad signaling, though they are not the major genetic components of otosclerosis. The similar mechanism was also come up by a recent study, which showed the BMP2 and BMP4 genes expressed more in otosclerotic stapes tissues than the normal ones.
Renin-Angiotensin-Aldosterone System-Related Genes: Both ACE (ACE I/D) and AGT (AGT M235T) are genetic polymorphisms of the Renin-Angiotensin-Aldosterone (RAA) system, and were hypothesized to influence the process of otosclerosis. They were found to be risk factors of otosclerosis in a Caucasian population. However, in another study of a larger Belgian-Dutch population failed to replicate the findings. There was no histologic evidence for the RAA system to express more protein in otosclerotic stapes footplates. No correlation was found between ACE gene and otosclerosis in another study. Therefore, whether the ACE and AGT genes play a role in otosclerosis is unclear.
HLA system: The HLA system is an important part of immune system. Early studies found the relations of HLA system and otosclerosis. However, the results could not be duplicated in other similar studies. Since the early studies were aimed at the serotypes of HLA instead of genotypes, it is hard to distinguish whether the genes are associated with otosclerosis on earth. For several years ago, a study reported that the HLA antigens were different in otosclerosis patients and the healthy controls based on Tunisian population.
RELN: The RELN gene is on chromosome 7q22.1, and takes part in neuronal migration. The association of RELN gene and otosclerosis was firstly reported by the study where a Genome-Wide Association (GWA) was performed via 555,000 single-nucleotide polymorphisms (SNPs) in 2009. Further studies confirmed the same findings next year, which might offer a different explanation of the molecular mechanism in the pathogenesis of otosclerosis. However, several subsequent studies based on various SNPs failed to find the evidence for the association between the RELN gene and the disease. The only exception is that one SNP (rs39399) presented a significant relation to otosclerosis. Also, one of the transcripts of RELN gene, RELN-203, was detected to express only in the tissues of otosclerosis rather than normal tissues. A recent research performed a case-control association study in a Tunisian-North African population, including 183 unrelated otosclerosis patients and 177 healthy subjects, and showed SNPs rs39335, rs39350 and rs39374 in RELN were significant otosclerosis-associated [63].
OPG: Early study reported that OPG (osteoprotegerin) was produced by stromal cells and osteoblasts cells, and was an important regulator physiologically in osteoclast differentiation and function. OPG gene took a part in inhibiting remodeling of the bone within the otic capsule and maitaining the normal auditory function, based on the study which used osteoprotegrin knockout mice. The recent study of Tunisian-North African population mentioned before also showed SNPs rs2073618 in OPG was significant otosclerosis-associated.
SERPINF1: Mutations in SERPINF1 (Serpin Peptidase Inhibitor, Clade F) were found to be the basis for a recessive form of osteogenesis imperfecta, a connective tissue disorder. PEDF (Pigment epithelium-derived factor) potently takes part in the inhibition of angiogenesis, and can regulate bone density definitely. Its expression is influenced by multiple mutations found in SERPINF1 gene. Expression of SERPINF1-012 transcript was found to reduce in otosclerosis patients with or without SERPINF1 mutations via RT-PCR. The findings may reveal a common pathogenic pathway in the otosclerosis. However, it could not be replicated recently [64].
So the relationship between SERPINF1 gene and otosclerosis remains unclear.
VDR: Both calcium and vitamin D replacement therapy have showed hearing improvement in a few cases of otosclerosis, it was suggeated Vitamin D level in plasma was related to otosclerosis. In further study, a possible relationship between the otosclerosis and VDR (Vitamin D Receptor) gene was found. Four polymorphisms of VDR gene showed three of them (Taq I, Apa I and Bsm I) were significantly associated with otosclerosis by RT-PCR [65].
MEPE: MEPE can encode a matrix extracellular phosphoglycoprotein, and plays an important role in inhibiting bone mineralization and resorption, suppressing renal calcification, and regulating serum phosphate. By analyzing MEPE gene in 89 otosclerosis families, 1604 unrelated affected subjects, and 1538 controls without screening, nonsense variations and rare frameshift in the MEPE gene were correlated with familial otosclerosis cases, and increased in unrelated otosclerosis subjects [66]. It was hypothesized that MEPE gene was possibly a rare risk factor of high effect size for otosclerosis. Also, an ASARM (acidic serine aspartate-rich MEPE)-Associated motif and an osteoregulin domain containing an RGD (arginyl-glycyl-aspartic acid) motif, which played a role in bone homeostasis, have been identified. It provided a possible molecular explanation of the disease.
Parathyroid hormone-related receptor: PTH (Parathyroid hormone) is an alkaline single-stranded peptide hormone secreted by the main cells of the parathyroid gland. PTH plays a key role in regulating the metabolism of calcium and phosphorus in human body. An early study showed that alkaline phosphatase level in serum was higher in otosclerosis patients of long duration, although another study demonstrated that bone mineral content and bone mineral concentration were found to be normal in otosclerosis patients [67]. A lower stimulation of cAMP production triggered by PTH was related to the lower PTH-PTHrP receptor mRNA expression in pathological stapes of otosclerosis patients. It was supported that the abnormal bone turnover in otosclerosis may be due to the abnormal cellular response to PTH.
SUMMARY
In the disease-causing genes, eight genetic loci of monogenic forms (OTSC1-5,7,8,10) have be found by linkage analyse, respectively on different chromosome, while it remains controversial that those candidate genes are associated with otosclerosis. Recently, some loci known for other disease were brought to the research of otosclerosis such as SERPINF1 and MEPE [68-72]. Actually, the evidence of most candidate genes is not sufficient or even doubtable as many studies failed to duplicate the positive findings. Though many unknown genetic loci may be involved, it is still hard to illustrate the exact mechanisms of otosclerosis to date [73-76].
ACKNOWLEDGEMENTS
The study was accomplished by the authors only. Contributions of all authors are listed as: Study design: Cai L, Xu Y; Data collection: Cai L, Xu Y, Liu Y and Chen Q; Manuscript preparation: Cai L; Manuscript revising: Xu Y, Liu Y and Chen Q.
REFERENCES
- Joseph RB, Frazer JP. Otosclerosis incidence in Caucasians andJapanese. Arch Otolaryngol. 1964; 80: 256-262.
- Altmann F, Glasgold A, Macduff JP. The incidence of otosclerosis as related to race and sex. Ann Otol Rhinol Laryngol. 1967; 76: 377-392.
- Tato JM, Tato JM Jr. Otosclerosis and races. Ann Otol Rhinol Laryngol. 1967; 76: 1018-1025.
- Jensen KJ, Nielsen HE, Elbrond O, Hansen HH. Mineral content of skeletal bones in otosclerosis. Clin Otolaryngol Allied Sci. 1979; 4: 339-342.
- Gregoriadis S, Zervas J, Varletzidis E, Toubis M, Pantazopoulos P, Fessas P. HLA antigens and otosclerosis. A possible new genetic factor. Arch Otolaryngol. 1982; 108: 769-771.
- Pedersen U, Madsen M, Lamm LU, Elbrond O. HLA-A, -B, -C antigens in otosclerosis. J Laryngol Otol. 1983; 97: 1095-1097.
- Browning GG, Gatehouse S. Sensorineural hearing loss in stapedial otosclerosis. Ann Otol Rhinol Laryngol. 1984; 93: 13-16.
- Yoo TJ. Etiopathogenesis of otosclerosis: a hypothesis. Ann Otol Rhinol Laryngol. 1984; 93: 28-33.
- Brookes GB. Vitamin D deficiency and otosclerosis. Otolaryngol HeadNeck Surg. 1985; 93: 313-321.
- Dahlqvist A, Diamant H, Dahlqvist SR, Cedergren B. HLA antigens inpatients with otosclerosis. Acta Otolaryngol. 1985; 100: 33-35.
- Sørensen MS, Nielsen LP, Bretlau P, Jørgensen MB. The Role of Type II Collagen Autoimmunity in Otosclerosis Revisited. Acta Otolaryngol. 1988; 105: 242-247.
- Nibu K, Okuno T, Nomura Y, Matsuki K, Juji T. [HLA and otosclerosis].Nihon Jibiinkoka Gakkai Kaiho. 1990; 93: 606-610.
- Altermatt HJ, Gerber HA, Gaeng D, Muller C, Arnold W. Immunohistochemical findings in otosclerotic lesions. HNO. 1992; 40: 476-479.
- Ramsay HA, Linthicum FH. Mixed hearing loss in otosclerosis:indication for long-term follow-up. Am J Otol. 1994; 15: 536-539.
- Svatko LG, Ismagilov Sh M. [The significance of genetic markers inotosclerotic hearing loss]. Vestn Otorinolaringol. 1994; (5-6): 13-15.
- Bodo M, Venti G, Baroni T, Bellucci C, Giammarioli M, Donti E, et al. Phenotype of in vitro human otosclerotic cells and its modulation by TGF beta. Cell Mol Biol (Noisy-le-grand). 1995; 41: 1039-1049.
- Miyazawa T, Tago C, Ueda H, Niwa H, Yanagita N. HLA associations in otosclerosis in Japanese patients. Eur Arch Otorhinolaryngol. 1996; 253: 501-503.
- Lolov SR, Edrev GE, Kyurkchiev SD, Kehayov IR. Elevated autoantibodies in sera from otosclerotic patients are related to the disease duration. Acta Otolaryngol, 1998; 118: 375-380
- McKenna MJ, Kristiansen AG, Bartley ML, Rogus JJ, Haines JL. Association of COL1A1 and otosclerosis: evidence for a shared geneticetiology with mild osteogenesis imperfecta. Am J Otol. 1998; 19: 604- 610.
- Tomek MS, Brown MR, Mani SR, Ramesh A, Srisailapathy CR, Coucke P, et al. Localization of a gene for otosclerosis to chromosome 15q25-q26. Hum Mol Genet. 1998; 7: 285-290.
- Grayeli AB, Sterkers O, Roulleau P, Elbaz P, Ferrary E, Silve, C. Parathyroid hormone-parathyroid hormone-related peptide receptor expression and function in otosclerosis. Am J Physiol. 1999; 277: E1005-1012.
- Singhal SK, Mann SB, Datta U, Panda NK, Gupta AK. Genetic correlationin otosclerosis. Am J Otolaryngol. 1999: 20: 102-105.
- McKenna MJ, Kristiansen AG. The role of measles virus and hereditary in the development of otosclerosis. New frontiers in immunobiology. 2000; 51-56.
- Udagawa N, Takahashi N, Yasuda H, Mizuno A, Itoh K, Ueno Y, et al Osteoprotegerin produced by osteoblasts is an important regulator in osteoclast development and function. Endocrinology. 2000; 141: 3478-3484.
- Declau F, Van Spaendonck M, Timmermans JP, Michaels L, Liang J, Qiu JP, Van de Heyning P. Prevalence of otosclerosis in an unselected series of temporal bones. Otol Neurotol. 2001; 22: 596-602.
- Van Den Bogaert K, Govaerts PJ, Schatteman I, Brown MR, Caethoven G, Offeciers FE, et al. (2001) A second gene for otosclerosis, OTSC2, maps to chromosome 7q34-36. Am J Hum Genet. 2001; 68: 495-500.
- Chen W, Campbell CA, Green GE, Van den Bogaert K, Komodikis C, Manolidis LS, et al. Linkage of otosclerosis to a third locus (OTSC3) on human chromosome 6p21.3-22.3. Journal of Medical Genetics. 2002; 39: 473-477.
- McKenna MJ, Kristiansen AG, Tropitzsch AS. Similar COL1A1 expression in fibroblasts from some patients with clinical otosclerosis and those with type I osteogenesis imperfecta. Ann Otol Rhinol Laryngol. 2002; 111: 184-189.
- McKenna MJ, Nguyen-Huynh AT, Kristiansen AG. Association of otosclerosis with Sp1 binding site polymorphism in COL1A1 gene: evidence for a shared genetic etiology with osteoporosis. Otol Neurotol. 2004; 25: 447-450.
- Rodriguez L, Rodriguez S, Hermida J, Frade C, Sande E, Visedo G, et al. Proposed association between the COL1A1 and COL1A2 genes and otosclerosis is not supported by a case-control study in Spain. Am J Med Genet A. 2004; 128a(1): 19-22.
- Van Den Bogaert K, De Leenheer EM, Chen W, Lee Y, Nurnberg P, Pennings RJ, et al. A fifth locus for otosclerosis, OTSC5, maps to chromosome 3q22-24. J Med Genet. 2004; 41: 450-453.
- Janssens K, ten Dijke P, Janssens S, Van Hul W. Transforming growth factor-beta1 to the bone. Endocr Rev. 2005; 26: 743-774.
- Brownstein Z, Goldfarb A, Levi H, Frydman M, Avraham KB. Chromosomal mapping and phenotypic characterization of hereditary otosclerosis linked to the OTSC4 locus. Arch Otolaryngol Head Neck Surg. 2006; 132: 416-424.
- Topsakal V, Fransen E, Schmerber S, Declau F, Yung M, Gordts F, et al. Audiometric analyses confirm a cochlear component, disproportional to age, in stapedial otosclerosis. Otol Neurotol. 2006; 27: 781-787.
- Zehnder AF, Kristiansen AG, Adams JC, Kujawa SG, Merchant SN, McKenna MJ. Osteoprotegrin knockout mice demonstrate abnormal remodeling of the otic capsule and progressive hearing loss. Laryngoscope. 2006; 116: 201-206.
- Chen W, Meyer NC, McKenna MJ, Pfister M, McBride DJ, Fukushima K, et al. Single-nucleotide polymorphisms in the COL1A1 regulatoryregions are associated with otosclerosis. Clin Genet. 2007; 71: 406-414.
- Declau F, Spaendonck MV, Timmermans JP, Michaels L, Liang J, Qiu JP, et al. Prevalence of histologic otosclerosis: an unbiased temporal bone study in Caucasians. Adv Otorhinolaryngol. 2007; 65: 6-16.
- Thys M, Schrauwen I, Vanderstraeten K, Janssens K, Dieltjens N, Van Den Bogaert K, et al. The coding polymorphism T263I in TGF-beta1 is associated with otosclerosis in two independent populations. Hum Mol Genet. 2007; 16: 2021-2030.
- Thys M, Van den Bogaert K, Iliadou V, Vanderstraeten K, Dieltjens N, Schrauwen I, et al. A seventh locus for otosclerosis, OTSC7, maps to chromosome 6q13-16.1. European Journal of Human Genetics. 2007; 15: 362-368.
- Bel Hadj Ali I, Thys M, Beltaief N, Schrauwen I, Hilgert N, Vanderstraeten K, et al. A new locus for otosclerosis, OTSC8, maps to the pericentromeric region of chromosome 9. Hum Genet. 2008; 123: 267-272.
- Imauchi, Y, Jeunemaitre X, Boussion M, Ferrary E, Sterkers O, Grayeli AB. Relation between renin-angiotensin-aldosterone system and otosclerosis: a genetic association and in vitro study. Otol Neurotol. 2008; 29: 295-301.
- Schrauwen I, Thys M, Vanderstraeten K, Fransen E, Dieltjens N, Huyghe JR, et al. (2008). Association of bone morphogenetic proteins with otosclerosis. J Bone Miner Res. 2008; 23: 507-516.
- Schrauwen I, Ealy M, Huentelma MJ, Thys M, Homer N, Vanderstraeten K, et al. A genome-wide analysis identifies genetic variants in the RELN gene associated with otosclerosis. Am J Hum Genet. 2009; 84: 328-338.
- Schrauwen I, Thys M, Vanderstraeten K, Fransen E, Ealy M, Cremers CW,et al. No evidence for association between the renin-angiotensin- aldosterone system and otosclerosis in a large Belgian-Dutch population. Otol Neurotol. 2009; 30: 1079-1083.
- Thys M, Schrauwen I, Vanderstraeten K, Dieltjens N, Fransen E, Ealy M, et al. Detection of rare nonsynonymous variants in TGFB1 in otosclerosis patients. Ann Hum Genet. 2009; 73: 171-175.
- Thys M, Van Camp G. Genetics of otosclerosis. Otol Neurotol. 2009;30: 1021-1032.
- Khalfallah A, Schrauwen I, Mnaja M, Fransen E, Lahmar I, Ealy M, et al. Genetic variants in RELN are associated with otosclerosis in a non- European population from Tunisia. Ann Hum Genet. 2010; 74: 399- 405.
- Priyadarshi S, Panda KC, Panda AK, Ramchander PV. Lack of association between SNP rs3914132 of the RELN gene and otosclerosis in India. Genet Mol Res. 2010; 9: 1914-1920.
- Schrauwen I, Ealy M, Fransen E, Vanderstraeten K, Thys M, Meyer NC, et al. Genetic variants in the RELN gene are associated with otosclerosis in multiple European populations. Hum Genet. 2010; 127: 155-162.
- Schrauwen I, Van Camp G. The etiology of otosclerosis: a combinationof genes and environment. Laryngoscope. 2010; 120: 1195-1202.
- Khalfallah A, Schrauwen I, Mnejja M, HadjKacem H, Dhouib L, Mosrati MA, et al. Association of COL1A1 and TGFB1 polymorphisms with otosclerosis in a Tunisian population. Ann Hum Genet. 2011; 75: 598- 604.
- Schrauwen I, Weegerink NJ, Fransen E, Claes C, Pennings RJ, Cremers CW, et al. A new locus for otosclerosis, OTSC10, maps to chromosome 1q41-44. Clin Genet. 2011; 79: 495-497.
- Bel Hadj Ali I, Ben Saida A, Beltaief N, Namouchi I, Besbes G,Ghazoueni E, et al. HLA class I polymorphisms in Tunisian patients with otosclerosis. Ann Hum Biol. 2012; 39: 190-194.
- Csomor P, Liktor B, Liktor B, Sziklai I, Karosi T. No evidence for disturbed COL1A1 and A2 expression in otosclerosis. Eur Arch Otorhinolaryngol. 2012; 269: 2043-2051.
- Csomor P, Sziklai I, Karosi T. Controversies in RELN/reelin expression in otosclerosis. Eur Arch Otorhinolaryngol. 2012; 269: 431-440.
- Schrauwen I, Khalfallah A, Ealy M, Fransen E, Claes C, Huber A, et al. COL1A1 association and otosclerosis: a meta-analysis. Am J Med GenetA. 2012; 158: 1066-1070.
-
Ertugay OC, Ata P, Kalaycik Ertugay C, Kaya KS, Tatlipinar A, KulekciS. Association of COL1A1 polymorphism in Turkish patients withotosclerosis. Am J Otolaryngol. 2013; 34: 403406.
- Iossa S, Corvino V, Giannini P, Salvato R, Cavaliere M, Panetti M, et al. The rs39335 polymorphism of the RELN gene is not associated with otosclerosis in a southern Italian population. Acta Otorhinolaryngol Ital. 2013; 33: 320-323.
- Liktor B, Csomor P, Szasz CS, Sziklai I, Karosi T. No evidence for the expression of renin-angiotensin-aldosterone system in otosclerotic stapes footplates. Otol Neurotol. 2013; 34: 808-815.
- Priyadarshi S, Ray CS, Panda KC, Desai A, Nayak SR, Biswal NC, et al. Genetic association and gene expression profiles of TGFB1 and the contribution of TGFB1 to otosclerosis susceptibility. J Bone Miner Res. 2013; 28: 2490-2497.
- Yildirim YS, Apuhan T, Duzenli S, Arslan AO. Otosclerosis and vitamin D receptor gene polymorphism. Am J Otolaryngol. 2013; 34: 454-457.
- Ealy M, Meyer NC, Corchado JC, Schrauwen I, Bress A, Pfister M, et al. Rare variants in BMP2 and BMP4 found in otosclerosis patients reduce Smad signaling. Otol Neurotol. 2014; 35: 395-400.
- Sommen M, Van Camp G, Liktor B, Csomor P, Fransen E, Sziklai I, et al. Genetic association analysis in a clinically and histologically confirmed otosclerosis population confirms association with the TGFB1 gene but suggests an association of the RELN gene with a clinically indistinguishable otosclerosis-like phenotype. Otol Neurotol. 2014; 35: 1058-1064.
- Richard C, Doherty JK, Fayad JN, Cordero A, Linthicum FH. Identification of target proteins involved in cochlear otosclerosis. Otol Neurotol. 2015; 36: 923-931.
- Rudic M, Keogh I, Wagner R, Wilkinson E, Kiros N, Ferrary E, et al. Thepathophysiology of otosclerosis: Review of current research. Hear Res. 2015; 330: 51-56.
- Priyadarshi S, Hansdah K, Ray CS, Biswal NC, Ramchander PV. Otosclerosis Associated with a De Novo Mutation -832G > A in the TGFB1 Gene Promoter Causing a Decreased Expression Level. Sci Rep. 2016; 6: 29572.
- Ziff JL, Crompton M, Powell HR, Lavy JA, Aldren CP, Steel KP, et al. Mutations and altered expression of SERPINF1 in patients with familial otosclerosis. Hum Mol Genet. 2016; 25: 2393-2403.
- Liktor B, Hirschberg A, Karosi T. [Otosclerosis. 1st part: pathogenesis]. Orv Hetil. 2018; 159: 1215-1220.
- Mowat AJ, Crompton M, Ziff JL, Aldren CP, Lavy JA , Saeed SR, et al. Evidence of distinct RELN and TGFB1 genetic associations in familial and non-familial otosclerosis in a British population. Hum Genet. 2018; 137: 357-363.
- Rekha S, Ramalingam R, Parani M. Pedigree Analysis and Audiological Investigations of Otosclerosis: An Extended Family Based Study. J Audiol Otol. 2018; 22: 223-228.
- Schrauwen I, Valgaeren H, Tomas-Roca L, Sommen M, Altunoglu U, Wesdorp M, et al. Variants affecting diverse domains of MEPE are associated with two distinct bone disorders, a craniofacial bone defect and otosclerosis. Genet Med. 2019; 1: 1199-1208.
- Valgaeren H, Sommen M, Beyens M, Vandeweyer G, Schrauwen I, Schepers A, et al. Insufficient evidence for a role of SERPINF1 in otosclerosis. Mol Genet Genomics. 2019; 294: 1001-1006.
- Wichova H, Shew M, Staecker H. Utility of Perilymph microRNA Sampling for Identification of Active Gene Expression Pathways in Otosclerosis. Otol Neurotol. 2019; 40: 710-719.
- Bouzid A, Tekari A, Jbeli F, Chakroun A, Hansdah K, Souissi A, et al. Osteoprotegerin gene polymorphisms and otosclerosis: an additional genetic association study, multilocus interaction and meta-analysis. BMC Med Genet. 2020; 21: 122.
- Hansdah K, Singh N, Bouzid A, Priyadarshi S, Ray CS, Desai A, et al. Evaluation of the Genetic Association and mRNA Expression of the COL1A1, BMP2, and BMP4 Genes in the Development of Otosclerosis. Genet Test Mol Biomarkers. 2020; 24: 343-351.
- Zhang Y, Tang Q, Xue R, Zhu X, Yang H, Gao Z. Analysis of the Genetic Characteristics of a Chinese Family With Otosclerosis. Ear Nose ThroatJ. 2021; 100: 7745-7805.