Expression and Prognostic Analysis of CDKs in Breast Cancer
- 1. Shandong University of Traditional Chinese Medicine, China
- 2. Binzhou Institute of Active Health (Binzhou Low Altitude Economy Research Institute), China
Keywords
• Breast Cancer
• CDKs
• Kaplan-Meier Plotter Analysis
• Tumor Markers
Citation
Yang YX, Zhang GJ, Zhang YD (2025) Expression and Prognostic Analysis of CDKs in Breast Cancer. Ann Breast Cancer Res 9(1): 1031.
INTRODUCTION
The Cyclin-Dependent Kinase (CDK) family comprises approximately 20 serine/threonine kinases that play crucial roles in regulating fundamental cellular processes, including cell cycle progression, transcriptional regulation, and RNA splicing [1]. Emerging evidence demonstrates that dysregulation of CDK signaling pathways significantly contributes to tumorigenesis and malignant progression, positioning CDKs as promising therapeutic targets in oncology [2]. Based on their functional specialization, CDKs can be categorized into two distinct groups: cell cycle-regulating CDKs (CDK1/CDC2, CDK2, CDK3, CDK4, and CDK6) and transcriptional CDKs (CDK7-9, CDK11-13, CDK19-20/CCRKs) [3,4].
The cell cycle regulatory subgroup governs phase transitions through phosphorylation of key substrates, with CDK3 specifically mediating G0-G1 phase transition [3]. In contrast, transcriptional CDKs orchestrate gene expression by phosphorylating the C-Terminal Domain (CTD) of RNA polymerase II, thereby regulating transcriptional initiation, elongation, and termination [2]. The critical involvement of CDKs in maintaining cellular homeostasis and their frequent dysregulation in malignancies have generated substantial interest in developing CDK-targeted therapies [4].
Global cancer epidemiology data from the World Health Organization’s International Agency for Research on Cancer (IARC) reveals breast cancer as the most prevalent malignancy worldwide, accounting for 11.7% of all new cancer cases in 2020 [5]. This public health challenge is particularly pronounced in China, where breast cancer constitutes 12.2% of global incidence and 9.6% of worldwide mortality, representing the most common female malignancy [6]. The escalating disease burden underscores the urgent need for novel therapeutic strategies targeting critical molecular pathways such as CDK signaling. Building on the established epidemiological significance and molecular pathogenesis of CDK dysregulation in breast carcinogenesis, we conducted a comprehensive bioinformatics investigation to elucidate the clinical relevance of CDK family members in breast malignancies.
Based on online databases (Oncomine, GEPIA, KEGG, cBioPortal and ENCORI, etc.), the expression level, clinicopathological features, survival and prognosis of CDKs in breast cancer patients were analyzed. In addition, miRNAs regulating CDKs were also analyzed. This integrative analysis not only delineates the functional landscape of CDKs in breast cancer progression but also identifies potential therapeutic vulnerabilities for CDKtargeted intervention strategies.
MATERIALS AND METHODS
Oncomine Database Analysis
The Oncomine platform (https://www.oncomine.org) was utilized to compare mRNA expression levels of CDKs between breast tumor tissues and adjacent normal tissues. Differential expression analysis was performed with the following parameters: The p-value threshold was 0.05, the multiple change was 2, and the genes were in the top 5%.
GEPIA Data
GEPIA is based on TCGA and GTEx databases and can be used to analyze RNA expression in a variety of cancer and normal tissue samples. GEPIA was used for correlation analysis of CDKs in breast cancer.
Kaplan and Meier Plotter
It might be used to predict the impact of target genes on survival of patients with different cancer types. We used Kaplan-Meier Plotter to analyze the prognostic value of CDKs and their regulatory miRNAs in breast cancer.
CBioPortal Database
The invasive breast cancer database was selected (including 1084 samples). CDKs cancer genome map based on cBioPortal analysis and construction. Mutations, putative copy number changes in DNA copies, and mRNA expression (microarray) Z-scores associated with diploid samples of the genome map were selected for analysis.
KEGG Database
KEGG analyzes visible genes on CDKs cell cycle maps with multiple sub databases, including genomes, biochemical reactions, biochemical substances, diseases and drugs, and common PATHWAY information.
ENCORI database
ENCORI (http://starbase.sysu.edu.cn/panCancer.php) to find targeted CDKs micrornas, and regulate CDKs miRNA expression levels were determined.
RESULTS
CDKs Transcription Levels in Breast Cancer Patients
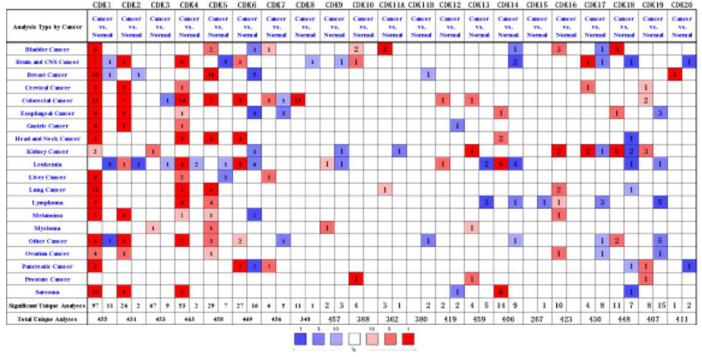
We used Oncomine database to and analyze the transcription levels of CDKs in clinical breast cancer samples and breast samples. In breast cancer patients, the mRNA expression levels of CDK1, CDK5 and CDK20 were significantly up-regulated, while the mRNA expression levels of CDK2, CDK6 and CDK11B were decreased, while the expression levels of other CDKs were not observed (Figure 1). Transcription levels of CDK1, CDK5 and CDK20 were significantly changed in breast cancer tumor tissues and normal tissues (Figure 1). Transcription levels of CDK1, CDK5 and CDK20 in different types of breast tumor tissues and normal tissues were significantly changed (Table 1). The absence or low expression of CDK10- CDK19 in breast cancer will not be analyzed in subsequent database analysis.
Figure 1: The transcriptional levels of CDKs in various cancers. The figure shows the number of datasets with higher expression levels of CDKs in various types of carcinoma samples compared to normal samples. The third row shows the expression of CDKs in breast cancer. Oncomine (http://www.oncomine.org).
Table 1: Transcriptional expression of CDKs in different types of breast cancer (Oncomine).
|
|
Type of Breast Cancer versus Normal Breast Tissue |
Fold Change |
P Value |
t Test |
Source and/or Reference |
|
CDK1 |
Mucinous breast carcinoma |
2.47 |
7.32 |
12.47 |
Curtis Breast Statistics |
|
Invasive ductal breast carcinoma |
3.27 |
4.98 |
45.05 |
Curtis Breast Statistics |
|
|
Medullary breast carcinoma |
4.03 |
3.78 |
13.15 |
Curtis Breast Statistics |
|
|
Breast carcinoma |
2.96 |
3.55 |
7.04 |
Curtis Breast Statistics |
|
|
Invasive ductal and invasive lobular breast carcinoma |
2.93 |
1.42 |
16.31 |
Curtis Breast Statistics |
|
|
Tubular breast carcinoma |
2.52 |
1.03 |
13.95 |
Curtis Breast Statistics |
|
|
Invasive breast carcinoma |
3.40 |
1.90 |
8.41 |
Curtis Breast Statistics |
|
|
Invasive lobular breast carcinoma |
2.33 |
2.36 |
18.72 |
Curtis Breast Statistics |
|
|
Mucinous breast carcinoma |
8.87 |
8.74 |
10.84 |
TCGABreast Statistics |
|
|
Male breast carcinoma |
7.00 |
1.09 |
13.62 |
TCGABreast Statistics |
|
|
Invasive ductal breast carcinoma |
5.86 |
1.51 |
25.54 |
TCGABreast Statistics |
|
|
Invasive lobular breast carcinoma |
4.76 |
8.14 |
10.61 |
TCGABreast Statistics |
|
|
Invasive breast carcinoma |
5.18 |
6.31 |
16.81 |
TCGABreast Statistics |
|
|
Intraductal cribriform breast adenocarcinoma |
2.05 |
8.25 |
8.46 |
TCGABreast Statistics |
|
|
CDK5 |
Lobular breast carcinoma |
2.45 |
3.03 |
11.11 |
Zhao Breast Statistics |
|
Invasive ductal breast carcinoma |
2.44 |
2.37 |
9.47 |
Zhao Breast Statistics |
|
|
Invasive breast carcinoma |
2.10 |
1.70 |
22.06 |
Gluck Breast Statistics |
|
|
Invasive ductal breast carcinoma |
2.17 |
1.85 |
21.51 |
TCGABreast Statistics |
|
|
Invasive ductal breast carcinoma |
2.16 |
1.73 |
41.10 |
Curtis Breast Statistics |
|
|
Tubular breast carcinoma |
2.01 |
1.27 |
18.93 |
Curtis Breast Statistics |
|
|
Invasive ductal and invasive lobular breast carcinoma |
2.11 |
9.17 |
21.98 |
Curtis Breast Statistics |
|
|
Mucinous breast carcinoma |
2.25 |
1.66 |
17.52 |
Curtis Breast Statistics |
|
|
Medullary breast carcinoma |
2.11 |
1.30 |
12.93 |
Curtis Breast Statistics |
|
|
Breast carcinoma |
2.02 |
1.22 |
13.64 |
Curtis Breast Statistics |
|
|
CDK20 |
Intraductal cribriform breast adenocarcinoma |
2.23 |
3.23 |
17.06 |
TCGABreast Statistics |
Transcriptional Level Analysis of CDKs in Breast Cancer
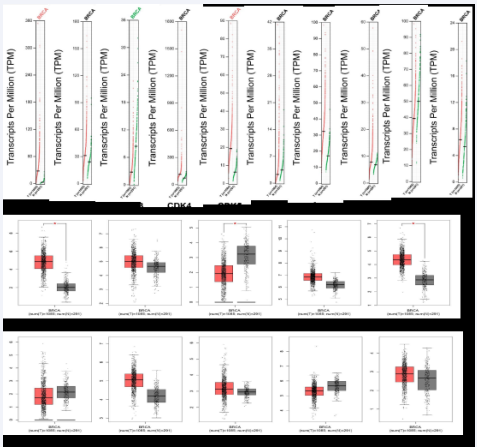
We compared CDKs gene expression in clinical breast cancer samples with normal breast tissue. The results showed that the expression levels of CDK1, CDK2, CDK4, CDK5, CDK7C, CDK8 and CDK20 in breast cancer tissues were higher than those in normal tissues, and there were significant differences in CDK1 and CDK5 (P < 0.05). The expression levels of CDK3 and CDK9 in breast cancer tissues were lower than those in normal breast tissues. CDK6 did not change (Figure 2).
Figure 2 The Expression of CDKs in Breast Cancer (A. scatter diagram; B. box plot). GEPIA database (http://gepia.cancer-pku.cn/).
Expression of CDKs in Breast Cancer Staging
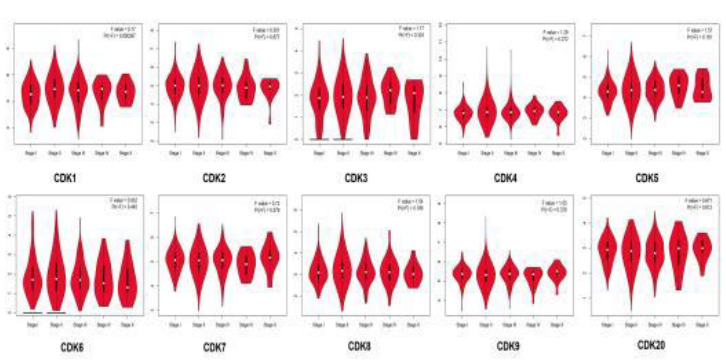
GEPIA database was used to analyze CDKs expression levels in breast cancer clinical samples at different stages. The results showed that CDK1 had significant difference in different stages of breast cancer (P < 0.05), while the other subtypes had no significant difference (Figure 3).
Figure 3 Correlation between CDKs expression and tumor stage in breast cancer patients. GEPIA database (http://gepia.cancer-pku.cn/).
Survival Rate and Prognostic Value of CDKs in Clinical Breast Cancer Patients
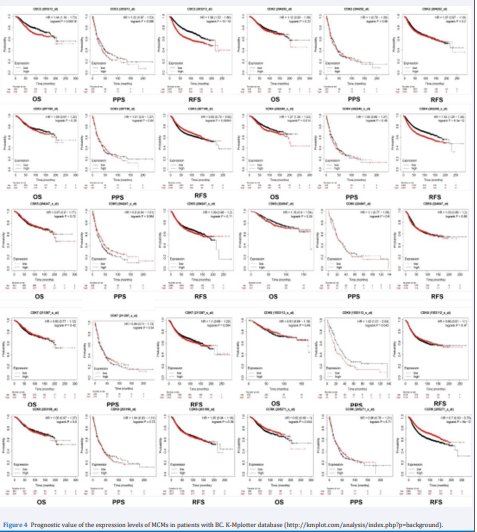
Kaplan-meier database was used to analyze the correlation between CDKs mRNA level and survival rate of breast cancer patients. The results showed that the recurrence free rate (RFS) and Overall Survival Rate (OS) of CDK1, CDK4 and CDK20 were significantly different (P < 0.05), the Recurrence Free Rate (RFS) of CDK3 was significantly different, and their mRNA levels were significantly correlated with RFS prolongation (P < 0.05).
These results suggest that CDK1, CDK3, CDK4 and CDK20 are associated with poor prognosis of breast cancer patients. The survival rate (PPS) after CDK8 progression was significantly different (P <0.05), and the mRNA level was correlated with the survival rate after CDK8 progression (Figure 4).
Figure 4 Prognostic value of the expression levels of MCMs in patients with BC. K-Mplotter database (http://kmplot.com/analysis/index.php?p=background).
Gene Recombination, Gene Correlation and CoExpression Gene Network of CDKs in Breast Cancer
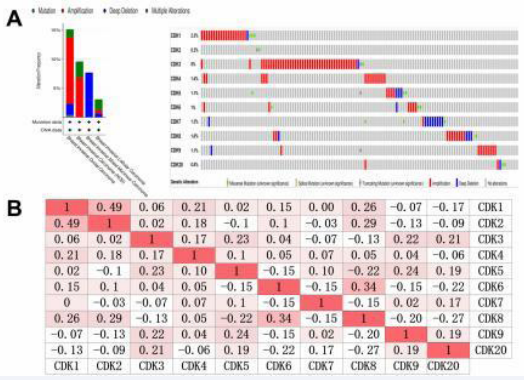
The cBioPortal database analyzed mutations or recombination of CDKs in breast cancer. As shown in Figure 5 (A), CDK was altered in 127 (13%) of 1084 breast cancer samples. In addition, the correlation of CDKs mRNA expression in breast cancer was analyzed. As shown in Figure 5 (B), There were significant positive correlations between CDK1 and CDK2, CDK4 or CDK6, CDK2 and CDK4 or CDK6 or CDK8, CDK3 and CDK4, CDK5 or CDK6, CDK4 and CDK5 or CDK7, CDK5 and CDK7, CDK6 or CDK8, CDK9 and CDK3, CDK5 or CDK20, CDK20 and CDK3, CDK5, CDK7 or CDK9 (R > 0, P < 0.05).
Figure 5 Analysis of alterations and correlation between members of CDKs in BC. (A) Gene expression and alteration analysis of CDKs in BC (cBioPortal). (B) Correlation analysis between different CDKs in BC. cBioportal (https://www.cbioportal.org/).
The Role of the Physicist CDKs in the Cell Cycle
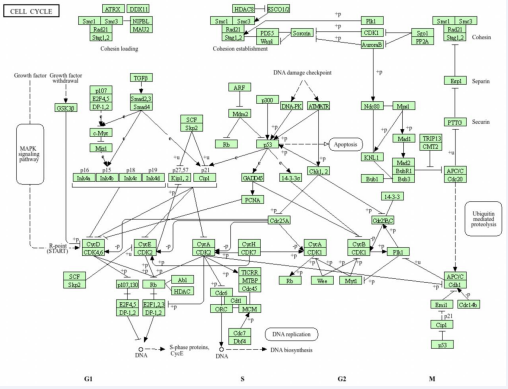
regulated by CDKs. As shown in (Figure 6), cyclin D is phosphorylated by CDK2, CDK4, and CDK6, driving cell cycle progression to G1. However, p16INK4a, p15INK4b, p18INK4c, and p19INK4d in the INK4 family interfere with CDK4 and CDK6 and prevent cyclin D phosphorylation and progression to G1. CDK2/Cycline E regulates cell cycle G1/S transition, followed by CDK2-Cycline A controlling S phase. CDK 1/Cycline A interferes with G2, and CDK1/Cycline B regulates G2/M transition and mitosis. The Kinase Inhibitor Protein (KIP) family, which includes p21CIP1, p27KIP1 and p57KIP2, binds to CDK and inactivates it. Binding of CDK2 to p27KIP1 at the G1 terminal delays the activation of the CDK2/Cycline E complex, leading to SKP1/SKP2 ubiquitination of CDK2 and destruction by proteasomes.
Figure 6 Visualization genes on cell cycle map. KEGG database (https://www.genome.jp/kegg/).
MicRNA Targeted CDKs
ENCORI database was used to screen miRNAs targeting CDKs. As shown in (Table 2), there were 42 CDK1-targeting miRNAs, among which 21 were negatively correlated;
Table 2: Analysis of miRNAs targeting CDKs.
|
|
Negative regulation number |
Positive regulation number |
The miRNA factor that targets the CDK gene |
|
CDK1 |
21 |
21 |
hsa-miR-329-3p, hsa-miR-410-3p, hsa-miR-944 |
|
CDK2 |
36 |
64 |
hsa-miR-29c-3p, hsa-miR-199a-5p, hsa-miR-664b-3p |
|
CDK3 |
1 |
1 |
hsa-miR-628-5p |
|
CDK4 |
20 |
31 |
hsa-miR-497-5p, hsa-miR-654-5p, hsa-miR-663a |
|
CDK5 |
3 |
6 |
hsa-miR-24-3p, hsa-miR-1287-5p, hsa-miR-1295a |
|
CDK6 |
94 |
274 |
hsa-miR-190b, hsa-miR-29c-3p, hsa-miR-375 |
|
CDK7 |
7 |
0 |
hsa-miR-139-5p, hsa-miR-362-5p, hsa-miR-500b-5p |
|
CDK8 |
35 |
78 |
hsa-miR-7b-5p, hsa-miR-30a-5p, hsa-miR-5691 |
|
CDK9 |
45 |
23 |
hsa-miR-23a-3p, hsa-miR-23b-3p, hsa-miR-455-3p |
|
CDK20 |
5 |
3 |
hsa-miR-24-3p, hsa-miR-574-3p, hsa-miR-4739 |
100 CDK2-targeting miRNAs, 36 negatively correlated; Two CDK3-targeting miRNAs, one negatively correlated; 51 CDK4-targeting miRNAs, and 20 were negatively correlated; Nine CDK5-targeting miRNAs, and three were negatively correlated; 368 CDK6-targeting miRNAs, 94 negatively correlated; 7 CDK7-targeting miRNAs, 7 negatively correlated 116 cdK8-targeting miRNAs, 35 of which were negatively correlated; There were 68 CDK9- targeting miRNAs, and 45 were negatively correlated; There were 8 CDK 20-targeting miRNAs, 5 of which were negatively correlated. Among them, CDK1-targeting miRNAs have significant relationships, such as CDK1-targeting HSA-Mir-329-3p (1.79), HSA-Mir-410-3p (7.66), HSA-Mir-944 (1.33), CDK2-targeting HSA-Mir-29C-3p (2.39), HSA-Mir-199a-5p (1.38), HSA-Mir-664b-3p (5.38), CDK3-targeting HSA-Mir-628-5p (2.23), CDK4-targeting HSA-Mir-497-5p (1.88), HSA-Mir-654-5p (1.05), HSAMir-663a (7.10), CDK5-targeting HSA-Mir-24-3p (1.23), HSA-Mir-1287-5p (6.32), HSA-Mir-1295a (3.94), CDK6- targeted HSA-Mir-190b (1.00), HSA-Mir-29C-3p (2.57), HSA-Mir-375 (1.75), CDK7-targeted HSA-Mir-139-5p (1.93), HSA-Mir-362-5p (1.66), HSA-Mir-500b-5p (9.98), Cdk8-targeting HSA-Mir-7b-5p (2.77), HSA-Mir-30a-5p (4.96), HSA-Mir-5691 (2.54), CDK9-targeted HSA-Mir23a-3p (9.14), HSA-Mir-23b-3p (2.17), HSA-Mir-455-3p (3.03), CDK20-targeting HSA-Mir-24-3p (1.87), HSA-Mir574-3p (7.05), HSA -Mir-4739 (1.46).
DISCUSSION
Our study investigated the expression levels, clinicopathological characteristics, survival rates, and prognostic value of CDKs in clinical breast cancer samples. This investigation aims to enhance our understanding of CDK functions and inform potential treatment strategies for breast cancer patients. CDK1, also known as CDC2 or CDC28A, is a pivotal regulator of cell cycle progression, controlling transitions through the G1/S and G2/M phases [7,8]. Dysregulation of CDK1 has been increasingly implicated in tumorigenesis, as its constitutive activation is closely associated with uncontrolled proliferation in various malignancies [9]. Notably, clinical studies have observed significant overexpression of CDK1 in ovarian epithelial carcinoma tissues, highlighting its potential as a diagnostic marker for early cancer detection [10].
Furthermore, in endometrial carcinoma, CDK1 expression positively correlates with tumor histological grade. Functional studies reveal that CDK1 promotes endometrial cancer cell proliferation, and its inhibition induces apoptosis and G2/M phase arrest [11]. Extending these findings to breast cancer, overexpression of CDK1 is consistently observed in breast cancer specimens, with a strong association between elevated CDK1 levels and poor clinical outcomes. This underscores its potential role as both a prognostic biomarker and a therapeutic target in breast oncology. CDK2, also referred to as CDKN2, is a crucial component in the regulation of the cell cycle, contributing to various biological processes including DNA damage response, intracellular transport, protein degradation, signal transduction, and the metabolism and translation of DNA and RNA [12].
Aberrant expression of CDK2 has been implicated in the pathogenesis of several cancers [13]. In gastric cancer, abnormal CDK2 expression is associated with enhanced malignancy and increased invasiveness of cancer cells. Studies have further demonstrated that CDK2 is aberrantly expressed in colon cancer, with significantly elevated levels in colon cancer cell lines such as HCT116, HCT15, and DLD-1 compared to normal colon epithelial cells. This upregulation of CDK2 is indicative of its role in facilitating the proliferation of gastric cancer cells and underscores its potential as a therapeutic target in the treatment of colon cancer [14]. Such findings highlight the critical role CDK2 plays in cancer biology, suggesting it as a promising candidate for targeted cancer therapy [15]. CDK3, a pivotal regulator of cell cycle progression, orchestrates transitions through the G0/G1 and G1/S phases, yet exhibits context-dependent roles in oncogenesis. While its dysregulated expression is widely documented across cancer cell lines, where it drives proliferation and malignant transformation [16], functional paradoxes emerge in specific tumor microenvironments. Notably, in primary non-metastatic breast tumors, aberrant CDK3 expression or overexpression suppresses migratory and invasive capacities, suggesting a tumor-restrictive role during early carcinogenesis [17]. Mechanistically, CDK3 phosphorylates NFAT3, forming a co-amplified signaling axis observed in cutaneous squamous cell carcinoma tissues, where CDK3, NFAT3, and phospho-NFAT3 levels significantly exceed those in adjacent normal tissues. This pathway may drive transcriptional reprogramming critical for tumor progression [18].
Conversely, in metastatic colorectal cancer, CDK3 overexpression correlates with aggressive phenotypes, facilitating Epithelial-Mesenchymal Transition (EMT) to enhance cellular motility, invasion, and distant metastasis [19]. These tissue-specific dichotomies underscore CDK3’s dual functionality in cancer biology, positioning it as a potential therapeutic target requiring context-dependent evaluation. CDK4 and CDK6, sharing 71% amino acid sequence homology, function as central regulators of G1 phase progression and G1/S transition through their interaction with D-type cyclins (D1, D2, D3) [2]. These kinases are frequently dysregulated in oncogenesis, with studies demonstrating abnormal activation and overexpression in ovarian cancer, where they drive uncontrolled cell cycle advancement [20]. In Non-Small Cell Lung Cancer (NSCLC), genomic alterations affecting CDK4/6-including amplifications and point mutations observed in 22-25% of patients-result in hyperactivation of proliferative pathways [21]. The clinical significance of CDK4/6 signaling is underscored by the FDA and EMA approval of selective CDK4/6 inhibitors for treating hormone receptor-positive, HER2-negative locally advanced or metastatic breast cancer in postmenopausal populations, marking a paradigm shift in targeted cancer therapy [22].
This regulatory landscape highlights CDK4/6 as both critical oncogenic drivers and actionable therapeutic targets across multiple malignancies. CDK7 serves as a master regulator of both cell cycle progression and transcriptional control, activating CDK1/2 while sustaining CDK4/6 activity to coordinate mitotic entry and transcriptional programs [26,27]. Its oncogenic role extends beyond cell cycle regulation, as CDK7 promotes tumorigenesis through transcriptional dysregulation of proliferative and survival pathways. Clinically, CDK7 overexpression correlates with increased incidence and poor prognosis in epithelial ovarian cancer [28]. In gastric cancer, elevated CDK7 expression drives tumor cell proliferation and associates with advanced histological grade and invasive depth [29]. These multifaceted roles underscore CDK7’s therapeutic potential, with selective inhibitors currently under investigation for precision oncology applications.
CDK8 orchestrates transcriptional regulation by phosphorylating the C-terminal domain of RNA polymerase II and modulates kinase activity through cyclin H phosphorylation to suppress CDK-Activating Kinase (CAK) function [30,31]. While frequently overexpressed in aggressive malignancies including colorectal and breast cancers-where its knockdown suppresses cancer cell proliferation, migration, and G0/G1 phase arrest [32- 34]-CDK8 exhibits paradoxical tumor-suppressive roles in endometrial cancer. Elevated CDK8 levels inversely correlate with tumor cell proliferation, invasion, and in vivo tumorigenesis in this context [35]. Notably, CDK8 inhibition demonstrates therapeutic efficacy in colon cancer models, effectively curbing malignant proliferation [33]. These context-dependent dualities position CDK8 as a compelling molecular target for precision oncology strategies in CDK8-driven cancers, though requiring tissue-specific therapeutic evaluation.
CDK9 was expressed throughout all phases of the cell cycle. CDK9 is a protein of potential clinical significance in tumors. CDK9 can bind cyclin T or K to form positive Transcription Extension Factor B (P-TEFB) [2]. CDK9 promotes genome integrity and plays a role in protecting against drugs that induce replication stress and DNA damage, and it has been proven to be dysregulated in many malignant tumors, including breast cancer [36,37], cervical cancer [38], etc. CDK9 inhibitors have also been shown to inhibit the growth of breast cancer cells and tumors [39-42]. CDK20 (Cell Cycle-Related Kinase, CCRK), a recently characterized member of the cyclin-dependent kinase family [43,44], exhibits conserved sequence homology with CDK7 and demonstrates novel CDK-Activating Kinase (CAK) activity critical for cell cycle regulation [45]. Emerging evidence underscores its oncogenic role across multiple malignancies, where CDK20 dysregulation drives tumorigenesis through proliferative signaling amplification.
In glioblastoma, CDK20 overexpression correlates with enhanced tumor aggressiveness and stemness maintenance [46], while in hepatocellular carcinoma, it promotes tumor progression via Wnt/βcatenin pathway activation [47-49]. Mechanistically, CDK20 knockdown suppresses cancer cell proliferation by arresting cell cycle progression and inducing apoptosis across diverse cancer models. These findings position CDK20 as a pivotal oncogenic driver and therapeutic vulnerability in CDK20-overexpressing tumors, warranting further exploration of targeted inhibition strategies.
CONCLUSION
This systematic investigation reveals critical dysregulation of CDK1, CDK5, and CDK20 in breast cancer, with elevated expression of CDK1, CDK4, CDK5, CDK7, CDK8, and CDK20 demonstrating clinical utility for patient stratification and molecular subtyping. These kinases exhibit strong correlations with advanced clinicopathological features and survival outcomes, supporting their dual roles as prognostic biomarkers and therapeutic targets. While Kaplan-Meier analyses highlight their prognostic potential, limitations in accounting for confounding variables necessitate validation through multivariate Cox regression models in future studies. Our findings establish CDKs as pivotal regulators of breast cancer progression and provide a framework for integrating CDK-driven signatures into precision oncology strategies to enhance prognostic accuracy and therapeutic decision-making.
REFERENCES
- Cicenas J, Valius M. The CDK inhibitors in cancer research and therapy. J Cancer Res Clin Oncol. 2011; 137: 1409-1418.
- Liao Y, Feng Y, Shen J, Hornicek FJ, Duan Z. The roles and therapeutic potential of cyclin-dependent kinases (CDKs) in sarcoma. Cancer Metastasis Rev. 2016; 35: 151-163.
- Carassou P, Meijer L, Le Moulec S, Aoun J, Bengrine-Lefèvre L. Le cycle cellulaire et ses cibles : inhibition des CDK [Cell cycle and molecular targets: CDK inhibition]. Bull Cancer. 2012; 99: 163-171.
- Chou J, Quigley DA, Robinson TM, Feng FY, Ashworth A. Transcription-Associated Cyclin-Dependent Kinases as Targets and Biomarkers for Cancer Therapy. Cancer Discov. 2020; 10: 351-370.
- Fahad Ullah M. Breast Cancer: Current Perspectives on the DiseaseStatus. Adv Exp Med Biol. 2019; 1152: 51-64.
- Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, et al.Breast cancer in China. Lancet Oncol. 2014; 15: e279-e289.
- Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007; 448: 811-815.
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009; 9: 153-166.
- Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div. 2010; 5: 11.
- Shi HR, Zhang RT. [Expression and significance of P53, P21WAF1 and CDK1 proteins in epithelial ovarian cancer]. Ai Zheng. 2009; 28: 882-885.
- Ying X, Che X, Wang J, Zou G, Yu Q, Zhang X. CDK1 serves as a novel therapeutic target for endometrioid endometrial cancer. J Cancer. 2021; 12: 2206-2215.
- Tadesse S, Anshabo AT, Portman N, Lim E, Tilley W, Caldon CE, et al. Targeting CDK2 in cancer: challenges and opportunities for therapy. Drug Discov Today. 2020; 25: 406-413.
- Tang Z, Li L, Tang Y, Xie D, Wu K, Wei W, et al. CDK2 positively regulates aerobic glycolysis by suppressing SIRT5 in gastric cancer. Cancer Sci. 2018; 109: 2590-2598.
- Lim TG, Lee SY, Huang Z, Lim DY, Chen H, Jung SK, et al. Curcumin suppresses proliferation of colon cancer cells by targeting CDK2. Cancer Prev Res (Phila). 2014; 7: 466-474.
- Liu FY, Wang LP, Wang Q, Han P, Zhuang WP, Li MJ, et al. miR-302b regulates cell cycles by targeting CDK2 via ERK signaling pathway in gastric cancer. Cancer Med. 2016; 5: 2302-2313.
- Wang L, Hu HY, Lin YL, Zhao ZX, Tan L, Yu P, et al. CDK3 expression and its clinical significance in human nasopharyngeal carcinoma. Mol Med Rep. 2014; 9: 2582-2586.
- Cao T, Xiao T, Huang G, Xu Y, Zhu JJ, Wang K, et al. CDK3, target of miR-4469, suppresses breast cancer metastasis via inhibiting Wnt/ β-catenin pathway. Oncotarget. 2017; 8: 84917-84927.
- Xiao T, Zhu JJ, Huang S, Peng C, He S, Du J, et al. Phosphorylation of NFAT3 by CDK3 induces cell transformation and promotes tumor growth in skin cancer. Oncogene. 2017; 36: 2835-2845.
- Lu J, Zhang ZL, Huang D, Tang N, Li Y, Peng Z, et al. Cdk3-promoted epithelial-mesenchymal transition through activating AP-1 is involved in colorectal cancer metastasis. Oncotarget. 2016; 7: 7012-7028.
- Zhang QF, Li J, Jiang K, Wang R, Ge JL, Yang H, et al. CDK4/6 inhibition promotes immune infiltration in ovarian cancer and synergizes with PD-1 blockade in a B cell-dependent manner. Theranostics. 2020; 10: 10619-10633.
- Qin Q, Ren Y, Zhong D. [Research Progress of CDK4/6 Inhibitors in Non-small Cell Lung Cancer]. Zhongguo Fei Ai Za Zhi. 2020; 23: 176- 181.
- de Groot AF, Kuijpers CJ, Kroep JR. CDK4/6 inhibition in early and metastatic breast cancer: A review. Cancer Treat Rev. 2017; 60: 130- 138.
- Contreras-Vallejos E, Utreras E, Gonzalez-Billault C. Going out of the brain: non-nervous system physiological and pathological functions of Cdk5. Cell Signal. 2012; 24: 44-52.
- Do PA, Lee CH. The Role of CDK5 in Tumours and TumourMicroenvironments. Cancers (Basel). 2020; 13: 101.
- Ehrlich SM, Liebl J, Ardelt MA, Lehr T, De Toni EN, Mayr D, et al. Targeting cyclin dependent kinase 5 in hepatocellular carcinoma--A novel therapeutic approach. J Hepatol. 2015; 63: 102-113.
- Zhang H, Christensen CL, Dries R, Oser MG, Deng J, Diskin B, et al. CDK7 Inhibition Potentiates Genome Instability Triggering Anti- tumor Immunity in Small Cell Lung Cancer. Cancer Cell. 2020; 37: 37-54.
- Schachter MM, Merrick KA, Larochelle S, Hirschi A, Zhang C, Shokat KM, et al. A Cdk7-Cdk4 T-loop phosphorylation cascade promotes G1 progression. Mol Cell. 2013; 50: 250-260.
- Kim J, Cho YJ, Ryu JY, Hwang I, Han HD, Ahn HJ, et al. CDK7 is a reliable prognostic factor and novel therapeutic target in epithelial ovarian cancer. Gynecol Oncol. 2020; 156: 211-221.
- Wang Q, Li M, Zhang X, Huang H, Huang J, Ke J, et al. Upregulation of CDK7 in gastric cancer cell promotes tumor cell proliferation and predicts poor prognosis. Exp Mol Pathol. 2016; 100: 514-521.
- Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulatedby cdk8-containing mediator complexes. Nature. 2000; 407: 102-106.
- Krempler A, Kartarius S, Gunther J, Montenarh M. Cyclin H is targeted to the nucleus by C-terminal nuclear localization sequences. Cell Mol Life Sci. 2005; 62: 1379-1387.
- Li XY, Luo QF, Wei CK, Deng-Feng L, Fang L. siRNA-mediated silencing of CDK8 inhibits proliferation and growth in breast cancer cells. Int J Clin Exp Pathol. 2014; 7: 92-100.
- Firestein R, Bass AJ, Kim SY, Ian FD, Serena JS, Guney I, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008; 455: 547-551.
- Firestein R, Shima K, Nosho K, Natsumi I, Yoshifumi B, Emeric B, et al. CDK8 expression in 470 colorectal cancers in relation to beta- catenin activation, other molecular alterations and patient survival. Int J Cancer. 2010; 126: 2863-2873.
- Gu W, Wang C, Li W, Fu-Ning H, Lifeng T, Jie Z et al. Tumor- suppressive effects of CDK8 in endometrial cancer cells. Cell Cycle, 2013; 12: 987-999.
- Cerami E, Gao J, Dogrusoz U, Benjamin EG, Selcuk OS, Bülent AA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2: 401-404.
- Gao J, Aksoy BA, Dogrusoz U, Gideon D, Benjamin G, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. SciSignal.2013;6:11.
- Liu X, Song J, Zhang Y, Huiquan W, Hongzhi S, Xiaomin F, et al. ASF1B promotes cervical cancer progression through stabilization of CDK9. Cell Death Dis. 2020; 11: 705.
- Li Y, Guo Q, Zhang C, Huang Z, Wang T, Wang X, et al. Discovery of a highly potent, selective and novel CDK9 inhibitor as an anticancer drug candidate. Bioorg Med Chem Lett. 2017; 27: 3231-3237.
- Mitra, P, Mitra P, Yang RM, Ramsay RG, Thomas JG. CDK9 inhibitors selectively target estrogen receptor-positive breast cancer cells through combined inhibition of MYB and MCL-1 expression. Oncotarget. 2016; 7: 9069-9083.
- Rajput S, Khera N, Guo Z, Jeremy H, Shunqiang L, Cynthia XM. Inhibition of cyclin dependent kinase 9 by dinaciclib suppresses cyclin B1 expression and tumor growth in triple negative breast cancer. Oncotarget. 2016; 7: 56864-56875.
- Sengupta S, Biarnes MC, Jordan VC. Cyclin dependent kinase-9 mediated transcriptional de-regulation of cMYC as a critical determinant of endocrine-therapy resistance in breast cancers. Breast Cancer Res Treat. 2014; 143: 113-124.
- Malumbres M, Harlow E, Hunt T, Hunter T, Jill ML, Gerard M, et al.Cyclin-dependent kinases: a family portrait. Nat Cell Biol. 2009; 11: 1275-1276.
- Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014; 15:122.
- Liu Y, Wu C, Galaktionov K. p42 a novel cyclin-dependent kinase- activating kinase in mammalian cells. J Biol Chem. 2004; 279: 4507- 4514.
- Ng SS, Cheung YT, An XM, Chen YC, Ming L, Gloria Hoi-Yee L, et al. Cell cycle-related kinase: a novel candidate oncogene in human glioblastoma. J Natl Cancer Inst. 2007; 99: 936-948.
- Feng H, Cheng AS, Tsang DP, May SL, Minnie YG, Yue SC, et al. Cell cycle-related kinase is a direct androgen receptor-regulated gene that drives beta-catenin/T cell factor-dependent hepatocarcinogenesis. J Clin Invest. 2011; 121: 3159-3175.
- Heitz PU, von Herbay G, Kloppel G, Komminoth P, Kasper M, Höfler H, et al. The expression of subunits of human chorionic gonadotropin (hCG) by nontrophoblastic, nonendocrine, and endocrine tumors. Am J Clin Pathol. 1987; 88: 467-472.
- Feng H, Yu Z, Tian Y, Ying-Ying L, May SL, Minnie YYG et al. A CCRK-EZH2 epigenetic circuitry drives hepatocarcinogenesis and associates with tumor recurrence and poor survival of patients. J Hepatol. 2015; 62: 1100-1111.