An Innovative AI-Powered Digital Pathology Information System
- 1. Department of Computational Pathology, NovinoAI, Fort Lauderdale, USA
- 2. Department of Veterans Affairs New York Harbor Healthcare System, USA
Abstract
Laboratory Information Systems, Image Management Systems, and Artificial Intelligence each have advanced pathology workflows, but their integration into a unified platform remains limited. We present FlexLIS, a cloud-based system that combines LIS, IMS, and AI into a secure AWS-based platform designed for scalability, compatibility, and compliance with HIPAA and SOC 2 standards. It supports advanced AI tools for multimodal image and data analysis; enabling automatic structured reporting; and the development of large-scale foundation models. Its user interface includes accession, reporting, image management, and administration modules which streamline laboratory workflows while offering powerful image viewing, annotation, and AI-assisted decision support, all with seamless interoperability with major electronic health record systems. Embedded AI modules provide intelligent slide label reading, automated quality control of whole-slide images, tissue typing and cataloging, and diagnostic and prognostic support across multiple organ systems. In summary, the AI-enabled FlexLIS system provides an unprecedented opportunity to optimize pathology workflows, enhance efficiency, and minimize diagnostic and clerical errors, thereby strengthening both patient safety and laboratory quality assurance.
Keywords
• Laboratory Information Systems; Image Management Systems; Artificial Intelligence; FlexLIS
Citation
Sali R, Zhang V, Baba F, Zhu M, Zhang W, et al. (2025) An Innovative AI-Powered Digital Pathology Information System. Ann Clin Pathol 12(1): 1178.
INTRODUCTION
Laboratory Information Systems (LISs) emerged in the 1970s as computer technology advanced and laboratories sought efficient ways to manage and process increasing volumes of clinical and research data [1]. In the 1990s, LISs incorporated greater automation and enhanced support for regulatory compliance, helping laboratories meet standards such as Clinical Laboratory Improvement Amendment (CLIA). Modern LISs consist of complex, interrelated software systems and infrastructure that support a wide range of laboratory information-processing needs [2]. LISs function across all phases of patient testing, including specimen acquisition and test ordering, specimen processing and tracking, result analysis and interpretation, and report creation and distribution [3]. In addition, LISs support the management of statistical reporting, quality control and improvement, revenue cycle operations, and other data essential to laboratory functions [1]. In the early 2000s, faster and more affordable digital scanners became available, marking the emergence of digital pathology [4]. Once traditional glass slides are digitized into electronic images, they must be managed as part of the clinical workflow. Image Management Systems (IMSs) are software platforms that provide the ability to organize and access images using image metadata, patient information, or other characteristics that associate images into meaningful groups [5]. Although IMSs are considered part of LISs, integration with legacy LISs has not yet been fully streamlined. In the 2020s, artificial intelligence (AI) has rapidly advanced and been increasingly applied in digital pathology, driven by improvements in AI architecture design and computational power, as well as the availability of large-scale language and vision models [6]. Vision AI refers to the capability of AI models to recognize patterns from training images and corresponding reports and to apply learned models to new, previously unseen images to support tasks such as classification and prediction. Pathology AI has been applied in digital pathology to extract quantitative features from whole slide images [7]. Despite these advances, current AI models mainly focus on screening and diagnosis [8], and their limited integration into pathology workflows has hindered widespread clinical adoption. To overcome these challenges, we introduce FlexLIS, a cloud-based, integrated LIS/IMS/AI system that enables users to access images and generate reports anytime and anywhere [9]. FlexLIS includes a range of AI tools that provide quality control throughout the pathology specimen processing and reporting workflow, improving laboratory efficiency and supporting pathologists in their daily tasks.
ARCHITECTURE AND USER INTERFACE DESIGN
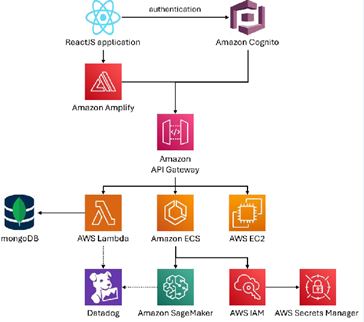
System Software Architecture: Amazon Web Services (AWS) is widely used in digital pathology and pathology AI [10]. FlexLIS is built with an API-first, microservices architecture on AWS. In practice, this means the platform is made up of many small, independent components (“microservices”), each packaged in a container—a lightweight, portable unit that makes them easy to deploy, update, and scale. All requests first pass through Amazon API Gateway, which acts as the secure entry point. It accepts encrypted HTTPS traffic, checks identities and credentials, validates requests, enforces limits, and then routes traffic to the right service.
For tasks that need an immediate response, the system uses Amazon Elastic Container Service (ECS), which automatically runs and scales containers while keeping them isolated for security. These containers are stored in Amazon Elastic Container Registry (ECR), a private repository that versions each build and scans it for vulnerabilities before use. For event-driven or scheduled jobs, such as processing files when they arrive, we rely on AWS Lambda, which runs code without the need to manage servers. Long-term data—including reports, images, exports, and machine learning features—is stored in Amazon S3, which offers encrypted storage, versioning to restore earlier file states, access controls, and lifecycle rules to archive or delete older data. All access runs over private connections inside our Virtual Private Cloud (VPC), a secure, isolated network environment. When machine learning is needed—for example, to flag quality issues or make triage suggestions—our services call Amazon SageMaker endpoints, which securely host and scale our trained models.
Security and monitoring are built into every layer. Amazon Inspector continuously scans software images (before deployment) and running workloads (after deployment) for known vulnerabilities, prioritizing fixes and feeding them into our release process. AWS Identity and Access Management (IAM) ensures every service only has the minimum permissions it needs, while sensitive information such as passwords and keys are encrypted in AWS Secrets Manager or Systems Manager Parameter Store with automatic rotation. All data flows are encrypted with TLS. For visibility, we use Datadog: it traces how requests move between services, captures logs, and measures the real user experience such as page load times and errors (with personal information scrubbed). These
insights are pulled together into dashboards, alerts, and incident timelines that give us a clear view of performance and reliability.
Software delivery is automated through a CI/CD pipeline. Code is built and tested, container images are pushed to ECR, vulnerability scans must pass before release, and deployments are rolled out either gradually (rolling updates) or in full swaps (blue/green releases). All infrastructure is defined as code (IaC), meaning every change is documented, auditable, and repeatable. Collectively, these practices support compliance with SOC 2 Trust Services Criteria (security, availability, confidentiality) and align with the NIST Cybersecurity Framework, covering all phases: Identify (asset inventories through IaC and automated scans), Protect (encryption, access controls, secrets management, and network segmentation), Detect (monitoring and alerts through Datadog), Respond (runbooks, on-call rotations, and incident timelines), and Recover (versioning, tested restores, and automated redeployment). Evidence such as gateway configurations, encryption policies, scan reports, deployment logs, and monitoring dashboards is maintained for auditing. (Figure 1).
Figure 1 Schematic overview of FlexLIS’s API-first microservices architecture on AWS
AI Engine Architecture: FlexLIS’s AI engine is conceived as an integral element of the entire workflow rather than as a peripheral add-on. Advanced AI models are seamlessly embedded across all critical stages, beginning with label interpretation and data ingestion, extending through slide quality assessment, diagnostic and prognostic analysis, and continuing to data validation and final report verification. This end-to-end integration
ensures that artificial intelligence consistently contributes to accuracy, efficiency, and reliability throughout the process, reinforcing both clinical and operational excellence.
The AI component of FlexLIS is designed with a flexible and modular architecture that supports a wide spectrum of model types, ranging from Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs) to emerging large-scale foundation models [11]. This diversity ensures that the system can accommodate a variety of analytical approaches and adapt to the rapid evolution of machine learning techniques in medical and computational pathology [12]. Users are able to take advantage of pre-trained models that have been integrated into the platform, providing immediate value and accelerating deployment. At the same time, the architecture allows users to train their own models directly within the system, enabling the creation of customized solutions tailored to specific datasets, institutional practices, or research needs. This dual pathway balances convenience with flexibility, offering both ready-to-use intelligence and an environment for innovation.
A central element of the design is the incorporation of active learning [13], which transforms routine use of FlexLIS into an opportunity for continuous model improvement. Pathologists remain in the loop during the production workflow, reviewing AI-generated outputs and providing feedback that is automatically captured by the system. This feedback is not limited to a single type of annotation but can operate at multiple levels of granularity depending on the nature of the AI task. For some applications, an entire slide image can be assigned a label, while in other cases a particular region may be annotated to reflect localized features of diagnostic importance. In more fine-grained scenarios, pathologists can annotate individual objects, such as cells or sub-cellular structures, creating highly detailed training data and refining the model’s performance in object-level recognition tasks. This layered approach to annotation makes the system equally well-suited for broad classification problems, region-specific analyses, and precise object detection.
Beyond image analysis, the architecture has been developed to integrate heterogeneous data streams from across the healthcare ecosystem. The system brings together digital pathology images, structured and unstructured clinical reports, laboratory results, and information from electronic health and medical record systems. By aligning these diverse data sources within a single framework, FlexLIS enables the training and deployment of advanced multimodal AI models capable
of capturing complex relationships between visual, textual, and numerical data. Such multimodal integration allows for more holistic and generalizable models that are better aligned with real-world clinical decision-making, supporting both diagnostic, prognostic and predictive applications.
Performance optimization and continuous innovation are central to FlexLIS. AI models are fine-tuned for speed and accuracy to support real-time or near–real-time inference, while the system functions as a living ecosystem in which new models can be seamlessly integrated as advances emerge. By combining model diversity, active learning, multimodal integration, and user-driven customization, the system establishes a robust foundation for precision medicine, enabling AI models to continuously adapt and improve while empowering customers to extend the platform to meet evolving clinical and research needs. More specific details about the AI applications supported by the system are provided in section 3.
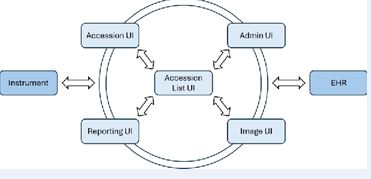
User Interface Architecture: To streamline clinical laboratory user workflow, FlexLIS is designed to have 4 UI modules for different functionalities and laboratory tasks (Figure 2). The accession UI is for entering specimen and patient demographic information. The reporting UI provides functions that pathologists enter diagnosis and release reports. The image management UI allows viewing the images associated with a specific case. The administration UI allows one to set up user privileges, templates (grossing, tests, reporting, etc), interface, etc (Figure 2).
Figure 2 Interaction among four UIs in FlexLIS and connectivity with EHR and laboratory instruments.
The Accession List UI is the center of FlexLIS which allows users to access other UIs depending on the assigned roles. It allows users to have an overview of all cases registered in FlexLIS. Depending on assigned roles, users can only perform certain tasks. For example, laboratory users can see the entire list of the cases and view details of each case. On the other hand, an office user can only see the cases belonging to the practice or provider and only be able
to open the final report as a PDF which can be downloaded or printed. When a user logs in as a pathologist, he can see the cases assigned to him and status of the cases, i.e. whether the case is ready to review, how many parts and slides, what stains performed, etc. The search filters allow users to search different fields including specimen types, tests performed, the date of image scanned, and pathologist’s notes etc.
The laboratory user can go to the Accession Detail UI by clicking Add New Accession (to add new cases) or click edit icon (to edit an existing case). In this UI, laboratory staff can enter patients’ demographic information including patient name, date of birth; specimen information including specimen type and number; and specimen details including site, measurements and tests. The requisition forms and other documents can be uploaded to the system and linked to each case for storage and retrieval. The laboratory staff can also see slide information and open the image window by clicking Open in Slide Viewer. The quality of the images including blurry (% of blurry), missing tissue and missing slides (how many slides) are flagged for each case in the accession list or in the slide image section where staff can address those images with issue before pathologists see the case.
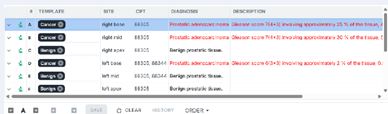
The Reporting UI (Figure 3), is designed for pathologists to enter the diagnosis of each specimen in a case. In the Reporting UI, pathologists can see patient and case information including clinical history, impression as well as the requisition form which are important for clinical-pathology correlation. The pathologist can enter diagnosis, comments, note and case comments which will be presented in the final reports. The diagnosis, comments and note can be entered by either pre-established codes or free writing. Pathologists can create their own templates/codes for diagnosis, comment or note. The diagnosis, comment and note can be color-coded based on the clinical importance so that providers can see the important diagnosis and comment easily. For example, cancer diagnosis is coded in red-color, dysplasia in orange, bacteria in blue, etc. Notably, preliminary diagnoses generated by AI after reviewing the images can also be auto-populated for each case. In addition, pathologists can
also add addendum or amendment as necessary. Finally, pathologists can review images associated with each case by clicking the image icon which will be shown in the Image Viewer UI. Pathologists can also see previous cases associated with a particular patient including reports and images by clicking History.
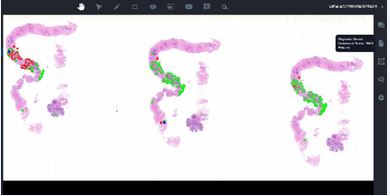
The most innovative UI is the Image Viewer (Figure 4), which allows access to the images associated with the cases. In this UI, all images including different stains are arranged in a tray format so that pathologists can see all images in alphabetical or sequential order. In addition, entire slides including labels can be seen by clicking the “eye icon”. FlexLIS provides an easy way for navigation and annotation of the images. The annotated images can be easily retrieved for later review. Users can set the zoom of the image at various magnifications for fast and easy view of the images. Furthermore, the user can directly insert an image into the report by selecting an area of the image. FlexLIS also offers easy ways for communication between pathologists and staff. Pathologists can email an image to a colleague for consultation or share the entire case with a colleague. This unique function makes the communication among pathologists easier and faster. Communication between pathologists and laboratory staff can be done via chat function. One unique feature is the direct link to ChatGTP where users can get more information regarding the pathology entity and differentiation diagnosis via text input or image input.
FlexLIS incorporates automated image analysis tools powered by AI to use pattern recognition algorithms to identify specific cell types, structures, or tissue changes, detect and delineate tumor boundaries or markers indicative of disease progression, and perform quantitative analysis such as cell counts and area measurements to aid in diagnosis or staging. Pathologists can annotate images with comments, markings or measurements, highlight areas on slides that require further review or analysis,
Figure 3 Reporting UI with diagnosis populated by prostate AI.
Figure 4 Image Viewer UI demonstrates prostate core biopsy with AI annotation.
marking tumor sizes, and adding text notes for sharing or future reference.
FlexLIS enables structured reporting, where findings from annotated images can be automatically populated into standardized reports (Figure 3). The seamless integration of AI between the IMS and LIS ensures that AI- generated assessments of images in IMS are automatically transcribed into pathology reports in LIS. This eliminates the need for manual data entry, allowing pathologists to focus on reviewing images in IMS and confirming diagnoses in LIS.
Security and Compliance: FlexLIS operates within a comprehensive information security program aligned with HIPAA, ISO 27001, and SOC 2 Type 2 requirements. The system boundary includes the LIS application, APIs, databases, and operational processes used to create, store, transmit, and audit Protected Health Information (PHI). We maintain an ISO-style Information Security Management System (ISMS) with defined assets, a living risk register, a Statement of Applicability, and recurring management reviews.
HIPAA safeguards—administrative, technical, and physical—are enforced through documented policies, workforce training, minimum-necessary data use, and role-based access. For SOC 2 Type 2, we evidence Security controls, with extensions for Availability and Confidentiality, through change tickets, access reviews, incident logs, and backup/restore testing conducted throughout the audit period.
To support compliance, we rely on AWS as our secure infrastructure backbone. Production workloads run in isolated VPCs across multiple Availability Zones, with centralized identity management via SSO and MFA, least- privilege IAM roles, and automated access reviews. PHI is encrypted both in transit (TLS) and at rest (AWS KMS with key rotation). Logging is centralized in S3 with Object Lock for immutability and lifecycle retention, while CloudTrail, Config, GuardDuty, Security Hub, and WAF provide continuous monitoring and threat detection. Backups are regularly validated with restore drills that measure RTO and RPO.
Our secure SDLC enforces gated code reviews, automated testing, container and dependency scanning (ECR, Inspector), signed releases, and auditable CI/CD change approvals. Non-production environments are configured to mask or exclude PHI by default.
The system also maintains built-in audit logs that
capture user access, image retrieval, AI model execution, and system changes, with automated alerts for anomalies. All AI algorithms undergo validation, version control, adversarial robustness testing, and ongoing performance drift monitoring. In addition, FlexLIS incorporates AI- driven tools that strengthen threat detection, encryption, access controls, and secure communications—integrating seamlessly with AWS native services to provide real- time detection, response, and evidence capture without disrupting clinical workflows.
This architecture enables FlexLIS to meet HIPAA safeguards, operate an ISO 27001-aligned ISMS, and maintain SOC 2 Type 2 attestation, while ensuring operational reliability. By combining AWS-native services with embedded auditability and AI-enhanced security controls, FlexLIS delivers reproducible assurances of security, availability, and confidentiality essential for regulated clinical workflows and dependable system operation.
Interface Capabilities: FlexLIS interfaces with analyzers, scanners, and middleware using HL7 v2.x over TCP with MLLP framing for real-time orders and results (Figure 2). Typical flows include ORM for orders, ORU for results, and optional ADT context. Connections are encrypted with TLS 1.2 or 1.3, with mutual TLS where supported; devices that lack native TLS can be fronted by a stunnel or a VPN. Delivery is verified with application ACKs, message control IDs, and replay-safe queues that prevent duplicates while supporting retries, connection pooling, and TCP keepalives for stability.
For batch or file-based devices, FlexLIS uses SFTP over SSH for secure exchanges of result files, logs, and images. Each instrument gets a chrooted drop folder, key-based authentication, least-privilege service accounts, and checksum verification. Idempotent parsers ingest from watchfolders into queue-backed pipelines with dead-letter handling for exceptions. All connectivity is gated by IP allow lists and audited end-to-end so every HL7 message or SFTP file is traceable to its accession, with monitoring on interface uptime, message latency, ACK success rate, SFTP acceptance rate, and alerts on orphan results or mapping errors.
FlexLIS is able to interface with a variety of Electronic Medical/Health Record (EHR) systems including hospital systems and outpatient practices. There are two types of EHR systems in office/outpatient practices, i.e. general practice and specialty. Most commonly used EHR systems in general practices include eClinical Works (eCW), which is widely used in outpatient practices and is strong in primary
care and multispecialty clinics. In addition, Athenahealth, MDland, and Practice Fusion are cloud-based systems and commonly used by smaller practices. Specialty-based EHR systems include Veradigm (urology), WRS Health (urology and gastroenterology), ModMed (gastroenterology), eCW- gastroenterology, Provation Apex (gastroenterology), etc.
FlexLIS is also able to interface with several enterprise EHR systems which are used in hospitals and large healthcare systems. Most commonly used enterprise EHR systems include Epic System (in U.S. hospitals and academic medical centers), Cerner (Oracle Health) (common in large hospital networks), and MEDITECH (often used in community hospitals). By combining these integrations with AI-powered cybersecurity, FlexLIS not only ensures interoperability but also protects against evolving digital threats, safeguarding the integrity and reliability of healthcare data across diverse environments. Furthermore, AI tools in FlexLIS ensure interfaces with EHR are secure and robust. AI tools can detect malfunctions of interface between EHR and other laboratory information systems and flag to staff so that interface issues can be addressed immediately.
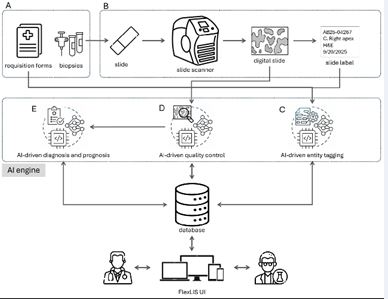
Figure 5 AI models are integral to FlexLIS. A. Requests arrive via requisition forms along with biopsies or samples. B. These requests are managed within lab operations. C. Requisition forms and slide labels are processed using AI-powered entity tagging to extract key information. D. Digital slide quality is assessed by AI to ensure low- quality slides do not affect downstream processes. E. Once quality is confirmed, slides are analyzed with AI models for both diagnostic and prognostic purposes.
AI-EMPOWERED WORKFLOW INTEGRATION
In recent years, the integration of Artificial Intelligence (AI) into digital pathology has transformed how diagnostic workflows are managed in clinical settings. FlexLIS is built with a strong foundation in computational pathology, offering a seamless combination of digital slide management and AI-powered analysis (Figure 5). Designed to assist pathologists throughout the diagnostic journey, this system addresses critical needs from slide quality control to automated diagnosis and prognosis, ensuring both accuracy and efficiency.
AI-Driven Specimen Documentation and Description: The first step in the specimen process at the laboratory is transferring information from the requisition form into the LIS, a step commonly referred to as accessioning. Although the LIS can interface with various EHR systems where test orders are placed, many requisitions still arrive at the laboratory in paper format. Manually entering patient and specimen information from paper requisitions into the LIS is often time-consuming, labor-intensive, and prone to errors. By leveraging Optical Character Recognition (OCR) and Large Language Models (LLMs) integrated into FlexLIS system, data from requisitions can be accurately extracted and stored in the database. The extracted information is then automatically populated into FlexLIS, significantly reducing staff effort while improving accuracy and efficiency.
Accurate documentation and description of specimens (i.e., grossing) received in the laboratory are essential for both diagnostic accuracy and legal protection. In FlexLIS workflow, each specimen is photographed from multiple angles, and the digital images are stored in FlexLIS, where they can be retrieved and reviewed by laboratory staff and pathologists as needed. An AI-powered vision model processes these images to automatically extract quantitative features, such as specimen measurements, and integrates them with information captured from the requisition to generate a comprehensive gross description. This AI-driven approach ensures specimen documentation that is more accurate, consistent, and seamless.
Intelligent Slide Label Reading and Data Extraction: Accurate and efficient extraction of metadata from histology and cytology slide labels is a critical component in automating laboratory workflows. These labels contain key identifiers such as patient demographics, accession numbers, and slide IDs, which are essential for downstream data linkage and traceability. Manual transcription is error-prone and inefficient, necessitating robust automated solutions.
FlexLIS employs state-of-the-art AI-powered OCR systems leveraging CNNs and transformer-based architectures to accurately detect and interpret textual and barcode information from slide labels. Preprocessing steps include image enhancement, noise reduction, and adaptive contrast adjustment to improve text visibility under
varying lighting and imaging conditions. The OCR model supports multi-format recognition including printed and handwritten text, as well as multi-language capabilities, enhancing robustness across diverse datasets.
After the OCR extracts text from slide labels, the system applies several advanced post-processing steps to maximize accuracy and reliability. First, confidence scoring [14], assigns a certainty level to each recognized element, helping to identify which parts may require further review. Next, syntactic validation rigorously checks that the extracted data follows expected formats and rules—ensuring, for example, that patient IDs or accession numbers conform to predefined patterns. Finally, context-aware error correction intelligently analyzes the surrounding information and common data patterns to automatically detect and correct likely mistakes, such as misread characters or swapped digits. Together, these features greatly reduce false positives and inconsistencies, resulting in highly trustworthy, clean data ready for downstream use. Extracted metadata is structured according to standardized schemas and transmitted securely to LIS and EHR using interoperable data exchange protocols such as HL7 and FHIR, enabling seamless integration within clinical workflows.
Designed for high throughput laboratory environments, the system architecture is highly scalable and efficient, supporting automated parallel processing of large volumes of slide images. This enables continuous ingestion and analysis with minimal human intervention, reducing operational bottlenecks and accelerating turnaround times. Containerized microservices orchestrated in cloud or on-premises infrastructure dynamically allocate computational resources to meet workload demands, ensuring responsiveness and fault tolerance.
The entire pipeline is developed in compliance with regulatory standards including HIPAA and GDPR, employing encryption at rest and in transit, role-based access controls, and audit logging to maintain data privacy and security. Continuous quality control is maintained through automated validation against reference datasets, human-in-the-loop verification for edge cases, and performance monitoring via detailed analytics dashboards. This comprehensive approach significantly improves data accuracy, optimizes laboratory efficiency, and enhances overall operational scalability.
AI-Driven Slide Quality Control: A critical upstream component of the digital pathology pipeline is the automated assessment of slide quality immediately following image acquisition and upload. Suboptimal Whole-
Slide Images (WSIs) — characterized by issues such as focal blur, incomplete tissue sections, or physical artifacts
— can significantly degrade the accuracy of downstream diagnostic algorithms and human interpretation, often necessitating repeat scans that delay case turnaround. To address this, FlexLIS system applies AI models such as CNNs and ViTs trained specifically to perform high-resolution Quality Control (QC) on digitized slides at the patch and slide levels. These models are optimized to detect a range of quality-degrading factors, including out-of-focus regions resulting from scanner miscalibration, tissue dropout, and acquisition artifacts such as staining irregularities. Once a predefined quality threshold is violated, the system automatically flags the affected WSIs and triggers alerts to FlexLIS for rescan workflows. This AI-driven QC module enables proactive mitigation of quality issues, reducing diagnostic uncertainty, minimizing manual review burden, and ensuring consistent, high-fidelity inputs for both human and AI-assisted pathology workflows.
AI-Driven Tissue Typing and Cataloging: Automated tissue classification and cataloging represent essential steps in both clinical and research-oriented digital pathology pipelines. Following WSI acquisition, FlexLIS leverages deep learning-based image classification models to perform organ-level tissue typing directly from high-resolution histopathological images. The outputs of these models are used to automatically assign WSIs to predefined anatomical categories—including, but not limited to, breast, lung, bladder, GI, prostate, skin, and cervical tissues—within the FlexLIS-integrated architecture. This automated stratification supports subspecialty-based diagnostic workflows by enabling case triage: for instance, WSIs identified as breast tissue are routed to breast pathology specialists, whereas thoracic cases are directed to pulmonary or thoracic pathologists. In translational and academic research settings, the same infrastructure facilitates cohort curation, allowing efficient aggregation of tissue samples by organ system (e.g., liver, kidney, lung) for biomarker discovery, AI models training, and population-based studies.
AI-Driven Diagnosis and Prognosis: One of the most transformative aspects of FlexLIS is its ability to provide automated diagnostic and prognostic support across a wide range of tissue types and disease conditions [9]. Trained on large, expertly annotated datasets, the AI models underpinning FlexLIS are capable of recognizing complex pathological patterns with a high degree of accuracy and consistency. For example, in prostate histopathology, the AI system assists pathologists by identifying cancerous regions within a slide, assessing their extent, and analyzing key architectural features of the tissue (Figure 3). It can
accurately assign Gleason grades based on glandular structures, quantify the size and proportion of malignant areas, and detect important morphological characteristics such as gland fusion, nuclear atypia, and mitotic activity [9]. These detailed, objective evaluations not only support diagnostic precision but also play a critical role in risk stratification and personalized treatment planning.
The same AI infrastructure has been adapted to analyze Gastrointestinal (GI) tissues, where it detects abnormalities throughout the upper and lower GI tract— including the esophagus, stomach, colon, rectum, and small intestine, such as the duodenum. In these tissues, the AI can identify a range of clinically relevant features, including inflammation, dysplasia, malignancy, and other histologic changes essential for accurate diagnosis and patient management.
In addition to histopathology, FlexLIS also supports cytology workflows [9]. It can evaluate cytological samples to detect and classify diverse cell types and subtle morphological abnormalities with high accuracy. This is particularly valuable in high-volume screening scenarios, such as Pap smears or effusion fluid analysis, where consistent recognition of fine cellular details is critical. By applying the same standards of precision and efficiency, the system contributes to timely and reliable diagnostic outcomes in cytopathology.
FlexLIS further enhances diagnostic workflows by intelligently prioritizing and supporting the tasks performed by pathologists [9]. When findings suggest significant pathology—such as cancer—the system automatically flags these cases for expedited review, ensuring that urgent conditions receive prompt attention. Moreover, in cases likely to require ancillary testing, the AI anticipates diagnostic needs and preorders immunohistochemical stains in advance. For example, it may initiate PIN4 staining for prostate biopsies or request
H. pylori stains for gastric samples based on morphological clues. This proactive, context-aware functionality reduces turnaround times, minimizes workflow interruptions, and improves overall laboratory efficiency—allowing pathologists to focus more on complex interpretive work and less on routine logistical steps.
AI-Driven Form and Data Validation: AI-driven form and data validation offers considerable potential to improve accuracy, efficiency, and reliability in both clinical and research environments [15]. Leveraging natural language processing and machine learning, such systems can automatically identify incomplete fields, inconsistent entries, and formatting errors in real time, thereby reducing manual oversight and minimizing downstream errors [16]. Unlike traditional rule-based approaches, AI models are able to learn contextual relationships within data—for example, detecting demographic outliers, cross- checking test results against expected biological ranges, and identifying discrepancies between structured and unstructured inputs—thereby ensuring greater data integrity. In addition, these systems can retrospectively analyze large volumes of historical data to uncover common patterns and trends, which can then be applied to guide future validation. When new entries deviate from established patterns, the system can proactively flag inconsistencies and alert users, facilitating early error correction. Collectively, these capabilities streamline workflows, enhance regulatory compliance, reduce the need for manual corrections, and support interoperability across diverse information systems such as LIS and EHRs.
LIMITATION AND FUTURE DIRECTION
Limitations
Like other pathology AI models, although FlexLIS provides advanced workflow automation, diagnostic support, and operational efficiency, several inherent limitations remain [6]. A primary challenge lies in data heterogeneity and integration. The FlexLIS platform processes data from a wide variety of instruments, scanners, and staining protocols, producing images and datasets with differing resolutions, color profiles, and formats. Similarly, clinical and molecular data are drawn from multiple EHR systems, external labs, and genomic sources, which can include inconsistent structures, missing fields, or incompatible terminologies. Although FlexLIS is designed to normalize and integrate these diverse inputs, gaps or inconsistencies can reduce the accuracy of AI- driven findings, limit the effectiveness of automated report generation, or require additional human oversight.
Another limitation concerns scalability and real- time processing. High-throughput laboratories generate large volumes of slides, images, and patient data daily. While our platform leverages optimized AI pipelines and agentic architecture, simultaneous processing of multiple multimodal datasets—especially when several AI agents interact in real time—can introduce latency in tasks such as anomaly detection, report drafting, or lab workflow monitoring. Computationally intensive operations, including large vision-language and LLM-based models, also demand substantial infrastructure, which may challenge smaller or resource-limited labs.
Interpretability and clinical trust present further constraints. Even with explainable AI features, complex models and LLM-generated outputs can be perceived
as “black boxes,” making it harder for pathologists and clinicians to fully understand the reasoning behind certain recommendations. Regulatory and compliance requirements add another layer of complexity: AI-driven analyses and report generation must remain fully auditable, validated, and reproducible. Continuous model updates and learning, while critical for performance improvement, must be carefully managed to avoid introducing errors or violating regulatory standards.
Operational integration also poses challenges. FlexLIS relies on smooth interfacing with laboratory instruments, sample tracking tools, and IT infrastructure. Variations in lab protocols, connectivity limitations, or network segmentation may temporarily hinder interoperability. While AI agents can augment workflow efficiency, monitor lab operations, and support decision-making, human expertise remains essential. Pathologists and lab personnel must validate critical cases, interpret ambiguous or rare findings, and make context-specific decisions that AI cannot fully replicate.
Finally, there are broader considerations related to bias, security, and workforce adaptation. Historical or skewed datasets could influence AI recommendations, particularly for rare diseases or underrepresented patient populations [17]. Secure handling of sensitive patient information across cloud or multi-institutional deployments is vital. Moreover, overreliance on automation could inadvertently reduce diagnostic practice among lab staff over time, emphasizing the need for ongoing human-AI collaboration and continuous training.
Collectively, these limitations illustrate that while FlexLIS can enhance efficiency, accuracy, and decision support, it remains an augmentation tool—pathologists and laboratory staff are indispensable for validation, complex interpretation, and patient-centered decision- making.
Future Directions
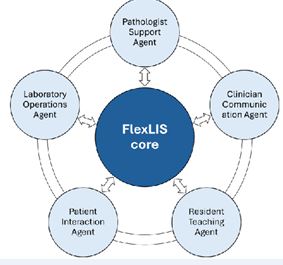
Figure 6 FlexLIS evolves into an ecosystem of specialized AI agents to enhance pathology, clinical care, education, and laboratory operations
The future development of FlexLIS is focused on evolving the platform into an ecosystem of integrated, specialized AI agents, each embedded throughout the workflow and designed to collaborate with specific stakeholders in pathology and healthcare (Figure 6) [18]. Central to this vision is the Pathologist Support Agent, which advances beyond our current capabilities of suggesting possible diagnoses, highlighting regions of interest, and sending findings to the report. The next- generation agent will deliver deep explanatory insights, interpreting morphological and cellular patterns, referencing established histological grading criteria and
immunohistochemical markers, identifying subtle or atypical tumor presentations, highlighting co-occurring features relevant to prognosis, and recommending additional stains or molecular tests. By integrating longitudinal patient data—including prior slides, genomic profiles, and treatment history—it will generate context- aware, evidence-linked recommendations. Furthermore, leveraging LLMs, the system will support automated generation of comprehensive, coherent pathology reports, synthesizing AI findings, molecular data, and clinical context to produce structured, clinically-ready documentation, thereby enhancing reproducibility, research-informed decision-making, and overall clinical efficiency.
Complementing this, the Clinician Communication Agent will translate complex pathology findings into actionable insights for treating physicians. It will not only summarize diagnoses but also provide risk stratification, prognostic interpretation, and therapeutic guidance, helping clinicians understand the implications of each finding in patient care. Education and training will be enhanced by the Resident Teaching Agent, which builds on AI-generated annotations and differential suggestions. Beyond visual guidance, it will provide interactive case- based learning, adaptive quizzes, progress tracking, and personalized feedback, effectively serving as a digital mentor for pathology trainees. The Patient Interaction Agent will improve communication with patients by delivering results in clear, empathetic language, explaining diagnoses, prognoses, and treatment options, and providing tailored educational materials. Meanwhile, the Laboratory Operations Agent—which already supports quality-control in current workflows—will further optimize lab operations by monitoring sample processing, predicting bottlenecks, enforcing standards, and offering proactive guidance for equipment management and supply chain coordination.
REFERENCES
- Henricks WH. Laboratory Information Systems. Clin Lab Med. 2016; 36: 1-11.
- Park SL, Pantanowitz L, Sharma G, Parwani AV. Anatomic pathology laboratory information systems: A review. Adv Anat Pathol. 2012; 19: 81-96.
- Yusof MM, Arifin A. Towards an evaluation framework for Laboratory Information Systems. J Infect Public Health. 2016; 9: 766-773.
- Cui M, Zhang DY. Artificial intelligence and computational pathology. Lab Invest. 2021; 101: 412-422.
- Zarella MD, Bowman D, Aeffner F, Farahani N, Xthona A, Absar SF, et al. A practical guide to whole slide imaging: A white paper from the digital pathology association. Arch Pathol Lab Med. 2019; 143: 222-234.
- Zhang DY, Venkat A, Khasawneh H, Sali R, Zhang V, Pei Z. Implementation of digital pathology and artificial intelligence in routine pathology practice. Lab Invest. 2024; 104: 102111.
- Baydoun A, Jia AY, Zaorsky NG, Kashani R, Rao S, Shoag JE, et al. Artificial intelligence applications in prostate cancer. Prostate Cancer Prostatic Dis. 2024; 27: 37-45.
- Zhu M, Sali R, Baba F, Khasawneh H, Ryndin M, Leveillee RJ, et al. Artificial intelligence in pathologic diagnosis, prognosis and prediction of prostate cancer. Am J Clin Exp Urol. 2024; 12: 200-215.
- Zhang DY, Sali R, Zhu M, Zhang V. Introduction of an Integrated Pathology Image Management, Artificial Intelligence, and Reporting System. J Vis Exp. 2025; 221.
- Bremer E, Kurc T, Gao Y, Saltz J, Almeida JS. Safe “cloudification” of large images through picker APIs. AMIA Annu Symp Proc.2016: 342- 351.
- Bilal M, Raza M, Altherwy Y, Alsuhaibani A, Abduljabbar A, Almarshad F, et al. Foundation models in computational pathology: A review of challenges, opportunities, and impact. arXiv preprint arXiv:250208333. 2025.
- Hosseini MS, Bejnordi BE, Trinh VQ, Chan L, Hasan D, Li X, et al. Computational pathology: A survey review and the way forward. J Pathol Inform. 2024; 15: 100357.
- Ren P, Xiao Y, Chang X, Huang P-Y, Li Z, Gupta BB, et al. A survey of deep active learning. ACM computing surveys (CSUR). 2021; 54: 1-40.
- Yang D, Tsai Y-HH, Yamada M. On verbalized confidence scores for llms. arXiv preprint arXiv:241214737. 2024.
- Kudo MS, Gomes de Souza VM, Estivallet CLN, de Amorim HA, Kim FJ, Leite KRM, et al. The value of artificial intelligence for detection and grading of prostate cancer in human prostatectomy specimens: A validation study. Patient Saf Surg. 2022; 16: 36.
- Golder S, Xu D, O’Connor K, Wang Y, Batra M, Hernandez GG. Leveraging natural language processing and machine learning methods for adverse drug event detection in electronic health/ medical records: A Scoping Review. Drug Saf. 2025; 48: 321-337.
- Alderman JE, Palmer J, Laws E, McCradden MD, Ordish J, Ghassemi M, et al. Tackling algorithmic bias and promoting transparency in health datasets: the STANDING Together consensus recommendations. Lancet Digit Health. 2025; 7: e64-e88.
- Zou J, Topol EJ. The rise of agentic AI teammates in medicine. Lancet. 2025; 405: 457.