Topographic Cell Cluster Sequencing Reveals Evolution Relationship and Driver Genes for Metastatic Invasive Micropapillary Carcinoma
- 1. Department of Laboratory Medicine, Third Affiliated Hospital of Zhengzhou University, China
- 2. Department of Breast Cancer Pathology and Research Laboratory, Tianjin Medical University Cancer Institute and Hospital, China
Citation
Shi Q, Fu L (2023) Topographic Cell Cluster Sequencing Reveals Evolution Relationship and Driver Genes for Metastatic Invasive Micropapillary Carcinoma. Ann Clin Pathol 10(1): 1164.
INTRODUCTION
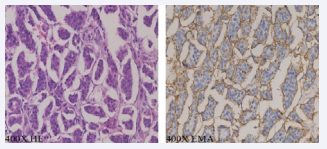
Invasive micropapillary carcinoma (IMPC) has very high rates of lymphovascular invasion and lymph node metastasis and has been reported in several organs [1-5]. Morphologically, IMPC tumor cells adhere to cell clustered structures with polarity reversed and located within empty stromal spaces, and tumor cells in recurrence, invasion, and metastasis are also arranged into cell cluster structures (Figure 1).
Figure 1: The IMPC tumor cells adhere to cell clustered structures with polarity reversed and located within empty stromal spaces.
Clinically, IMPC exhibits higher rates of lymphovascular invasion, lymph node metastasis, recurrence, and distant metastasis, and a poorer prognosis [6,7]. The unique clustered growth pattern and aggressive biological behaviors make IMPC a good model for studying the mechanism of breast cancer invasion and metastasis.
In the past few years, we have been committed to the mechanisms of high invasion and metastasis of breast IMPC since 1994 [6]. It was found that IMPC is not only in the primary tumor but also in the invaded lymphovascular and metastatic lymph nodes, the tumor cells are arranged in tumor cell clusters with reversed polarity. Therefore, we proposed a hypothesis that “IMPC tumor cells growth, invasion and metastasis with clustered arrangement”. A series of studies also revealed by our team successfully examined the pathology, clinical features, and metastatic mechanism of the “clustered metastasis of IMPC tumor cells” [8-11]. Recently our team revealed the transcriptome of IMPC and reveals its extensive heterogeneity employed by spatial transcriptomics sequencing [12]. We also revealed the mechanism of tumor cell cluster invasion and metastasis in IMPC of the breast performed by single-cell RNA sequencing (unpublished data). However, the genomic variants and evolutionary relationship of tumor cell clusters of IMPC not yet revealed.
To address this possibility, in our recent study published in Nature Communications, topographic cell cluster sequencing (TCCS) was used to reveal the genomic features and evolutionary relationship of IMPC tumor cell clusters [13]. TCCS is a new method combined with laser-capture microdissection (LCM), spatial information of cell clusters in tissue sections and DNA sequencing. This method not only obtains the sequencing data from tumor cell clusters of IMPC but also the spatial location information of tumor cell clusters in the tissue sections. It is also possible to study the evolution of the invasion and metastasis of IMPC tumor cell clusters from primary to lymph node metastases.
Our data demonstrated that compared to SNV, CNV played a stronger correlation with the potential of IMPC lymph node metastasis, thus supporting that CNV plays a more important role in the characteristic of IMPC high lymph node metastasis. This indicates that our focus on CNVs is the key to research the invasion and metastasis mechanism of IMPC. So, we further found that primary IMPC components have far more unique genes than IDC, for example, the copy-number loss of AKT3, CCND1, and gains of MYCN, genes were observed in IMPC. Furthermore, IMPC and IDC accumulate more specific CNVs during metastasis than primary tumors. Our team previously found that even if the proportion of IMPC component was small, the lymphovascular invasion and lymph node metastasis was higher than IDC, and the metastatic lymph nodes are mainly IMPC components [8].
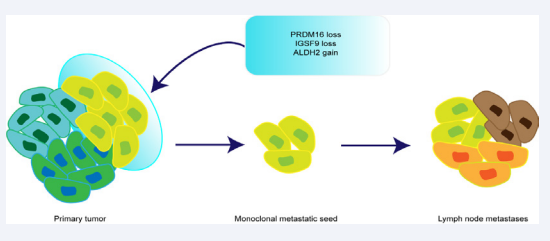
We constructed the evolutionary relationship of IMPC tumor cell clusters in the primary tumor, and found that the tumor cell clusters in the primary lesions were heterogeneous, and the cell clusters located inside the tissue sections were in the early stages of tumor evolution, and the cell clusters outside the tissue section were in the late stage of tumor evolution. This suggests that tumor cell clusters located at the edge of tumor tissue sections accumulate more CNV genes and have stronger invasion and metastasis capabilities. To further reveal the metastatic path of IMPC tumor cell clusters, we constructed a phylogenetic tree between tumor cell clusters from the primary tumor to metastatic lymph nodes. It was found that the cell clusters in metastatic foci originated from a single subclone in the primary tumor. This indicates that IMPC is invaded and metastasized using a monoclonal metastatic seed route. Further analysis of the genomic CNV characteristics of the monoclonal metastatic seed showed that the copy-number losses of PRDM16, IGSF9 genes and the copy-number gain of the ALDH2 gene were the highly frequency genomic CNV characteristics of IMPC monoclonal metastatic seed. This manifests that PRDM16, IGSF9, and ALDH2 genes are the key driving genes for IMPC high lymph node metastasis. Immunohistochemistry results of 86 IMPC patients also demonstrated that low expression of IGSF9 and PRDM16 and high expression of ALDH2 are significantly associated with lymph node metastasis of IMPC.
Overall, we found that genomic CNV plays a more critical role in the biological behavior of IMPC’s highly metastasis. In addition, IMPC evolves with monoclonal metastatic seed. Harboring the copy-number losses of PRDM16, IGSF9 genes, and the copynumber gain of the ALDH2 gene, tumor cell cluster will easier break away from the primary tumor, invade and metastases to lymph nodes (Figure 2).
Figure 2: The monoclonal metastatic seed harboring the copy-number losses of PRDM16, IGSF9 genes, and the copy-number gain of the ALDH2 gene in IMPC primary tumor
These three genes identified in this study provide key targets for precise diagnosis and treatment of IMPC patients.
Based on this least research result, Prof. Li Fu recently has led a team to jointly marker ALDH2 and the overexpressed folate receptor-α (FR-α) on the surface of tumor cells for in vivo visualization of pathological diagnosis research (Figure 3).
Figure 3: Schematic diagram of non-invasive pathological diagnosis of in vivo visualization of ALDH2 and folate receptor-α
Correlate the fluorescence imaging characteristics of ALDH2 and FR-α, the key molecules of breast cancer metastasis seed cell evolution, with tumor cell localization, establish a functional classification index system for invasive/metastatic breast cancer, and form a non-invasive pathological diagnosis of breast cancer and surgical navigation. New technology, dedicated to improving survival and quality of life for breast cancer patients.
REFERENCES
- Siriaunkgul S, Tavassoli FA. Invasive micropapillary carcinoma of the breast. Mod Pathol. 1993; 6: 660-2.
- Amin MB, Ro JY, el-Sharkawy T, Lee KM, Troncoso P, Silva EG, et al. Micropapillary variant of transitional cell carcinoma of the urinary bladder. Histologic pattern resembling ovarian papillary serous carcinoma. Am J Surg Pathol. 1994; 18: 1224-32.
- Sakamoto K, Watanabe M, De La Cruz C, Honda H, Ise H, Mitsui K, et al. Primary invasive micropapillary carcinoma of the colon. Histopathology. 2005; 47: 479- 84.
- Kuroda N, Hamaguchi N, Ohara M, Hirouchi T, Miyzaki E, Mizuno K. Intracytoplasmic lumina in invasive micropapillary carcinoma of the lung. Diagn Cytopathol. 2006; 34: 224-6.
- Kitagawa H, Nakamura M, Tani T, Tajima H, Nakagawara H, Ohnishi I, et al. A pure invasive micropapillary carcinoma of the pancreatic head: Long disease- free survival after pancreatoduodenectomy and adjuvant chemotherapy with gemcitabine. Pancreas. 2007; 35: 190-2.
- Fu L, Ikuo M, Fu XY, Liu TH, Shinichi T. [relationship between biologic behavior and morphologic features of invasive micropapillary carcinoma of the breast]. Zhonghua Bing Li Xue Za Zhi. 2004; 33: 21-5.
- Guo X, Chen L, Lang R, Fan Y, Zhang X, Fu L. Invasive micropapillary carcinoma of the breast: Association of pathologic features with lymph node metastasis. Am J Clin Pathol. 2006; 126: 740-6.
- Cui LF, Guo XJ, Wei J, Liu FF, Fan Y, Lang RG, et al. Overexpression of tnf-alpha and tnfrii in invasive micropapillary carcinoma of the breast: Clinicopathological correlations. Histopathology. 2008; 53: 381-8.
- Liu F, Lang R, Wei J, Fan Y, Cui L, Gu F, et al. Increased expression of sdf-1/cxcr4 is associated with lymph node metastasis of invasive micropapillary carcinoma of the breast. Histopathology. 2009; 54: 741-50.
- Li W, Liu F, Lei T, Xu X, Liu B, Cui L, et al. The clinicopathological significance of cd44+/cd24-/low and cd24+ tumor cells in invasive micropapillary carcinoma of the breast. Pathol Res Pract. 2010; 206: 828-34.
- Li S, Yang C, Zhai L, Zhang W, Yu J, Gu F, et al. Deep sequencing reveals small rna characterization of invasive micropapillary carcinomas of the breast. Breast Cancer Res Treat. 2012; 136: 77-87.
- Lv J, Shi Q, Han Y, Li W, Liu H, Zhang J, et al. Spatial transcriptomics reveals gene expression characteristics in invasive micropapillary carcinoma of the breast. Cell death & disease. 2021; 12: 1095.
- Shi Q, Shao K, Jia H, Cao B, Li W, Dong S, et al. Genomic alterations and evolution of cell clusters in metastatic invasive micropapillary carcinoma of the breast. Nature Communications. 2022; 13: 111.