Investigating the Role of Campylobacter jejuni Nickel-Binding Protein NikZ in Chemosensing
- 1. Institute for Glycomics, Griffith University, Australia
Abstract
NikZ has been characterised as the nickel binding protein in C. jejuni and is considered to play an important role in transferring nickel into the cell. In this study, we used the amino acid, glycan and small molecule arrays to investigatewhether NikZ is involved in C. jejuni chemotaxis indirectly. Protein-protein interaction analysis of NikZ with periplasmic domains of group a transducer like chemosensory proteins, (Tlps), showed no interactions, and NikZ was unable to bind any of the ligands on the arrays. Moreover, chemotaxis assays revealed that nickel is chemorepellent for both the C. jejuni strain NCTC 11168-O and its nikZ− isogenic mutant. All nickel concentrations examined in this study (0.1, 0.5, 1 and 2 mm) decreased the chemo directed motility of both, the wild type and nikZ− mutant strains. Presence of nickel decreased the growth rate of the wild type and the nikZ− mutant isogenic strains at all tested concentrations, except for 0.5 mm nickel, which restored the growth rate of the nikZ− mutant strain, similar to that of the wild type in absence of nickel. These data show that NikZ is not involved in C. jejuni chemosensing.
Citation
l-Kass A, Elgamoudi B, Day C, Korolik V (2018) Investigating the Role of Campylobacter jejuni Nickel-Binding Protein NikZ in Chemosensing. Ann Clin Med Microbiol 3(1): 1017.
Keywords
• Campylobacter jejuni; Chemotaxis; NikZ; Periplasmic
binding protein; Nickel binding protein
ABBREVIATIONS
Tlp: Transducer Like Protein; Ni-BP: Nickel Binding Protein
INTRODUCTION
Campylobacter jejuni is considered to be a leading cause of bacterial gastroenteritis in the world. It is suggested that the campylobacteria are responsible for 400-500 million cases of gastroenteritis annually [1,2]. C. jejuni is a microaerophilic bacteria and it needs nickel and iron to activate the [NiFe] hydrogenase to be able to tolerate toxic levels of O2 . The bacteria also depends on hydrogenase and formate dehydrogenase to provide a suitable conditions for growth [3]. Nickel is a heavy metal and it activates many important enzymes like [NiFe] hydrogenase, urease, methyl coenzyme M-reductase, CO dehydrogenase in many living organisms [4]. Although nickel is important for growth of bacterial cells, it is toxic at high concentrations [5]. For example, high levels of nickel are known to be inhibitory for Escherichia coli [6- 8]. While exact reason for this toxicity had not been established, many theories have been put forward to explain the mechanisms of nickel toxicity in the bacterial cells. These theories are: nickel has indirect role in oxidative stress, it replaces the heavy metal in the metalloproteins and inhibits enzymes by binding to catalytic sites of the enzymes [9].
In E. coli, aspartate/maltose chemoreceptor (Tar) senses nickel without interaction with nickel binding protein, NikA [10].
Nickel is sensed by TlpC in Helicobacter pylori and been shown to be a repellent [11,12]. Although nickel acts as a repellent, it is important for H. pylori survival as its paucity prevents maturation of hydrogenase, and subsequently, the colonization of the human host can be affected [13].
Similarly, nickel plays an important role in oxidative stress in C. jejuni NCTC 11168. Nickel and iron [NiFe] activate the hydrogenase leading to O2 tolerance via conversion to H2 O [14]. C. jejuni encodes an ATP cassette (ABC) transporter encoded by (nikZ YXWV) operon (cj1584c–cj1580c) [15]. NikZ protein shares similarity with NikA, the nickel binding protein in E. coli, and is encoded by the gene Cj1584c, which had been characterized as nickel binding protein, Ni-BP [14]. Considering that C. jejuni has a small genome with many proteins being of dual function [15], we investigated the potential role of nickel binding protein NikZ in the indirect sensing in chemotaxis in C. jejuni.
MATERIALS AND METHODS
Bacterial strains, growth conditions and plasmids
C. jejuni cells have been cultured on Colombia Blood Agar supplemented with antibiotics (Vancomycin 10 μg/mL, trimethoprim 2.5 μg/mL and polymexin B 0.3 μg/mL (Sigma Aldrich)) at micro aerobic conditions (5% O2 , 10% CO2 , 85% N2 ) at 42ºC. E. coli cells have been cultured on LB agar supplemented with 100 μg/ml of ampicillin (Sigma-Aldrich) at 37ºC. Bacterial strains and plasmids used in this study are listed in (Table 1).
Table 1: The percentage of nickel inhibition in t-HAP assay.
| Wild type | nikZ- mutant | ||||
| Cell numbers | Inhibition rate | Cell numbers | Inhibition rate | ||
| 0.1mM Nickel | 100mM Serine | 6.02 × 106 | 23.4 % | 2.3 × 106 | 48.4 % |
| 100mM Serine + 0.1mM Ni | 4.6 × 106 | 1.18 × 106 | |||
| 0.5mM Nickel | 100mM Serine | 7.9 × 106 | 51.9 % | 2.5 × 106 | 55.5 % |
| 100mM Serine + 0.5mM Ni | 3.8 × 106 | 1.1 × 106 | |||
| 1mM Nickel | 100mM Serine | 7.9 × 106 | 84.4 % | 4.01 × 106 | 78.6 % |
| 100mM Serine + 1mM Ni | 1.2 × 106 | 0.8 × 106 | |||
| 2mM Nickel | 100mM Serine | 10.7 × 106 | 88.2 % | 3.4 × 106 | 96.5 % |
Primer design and molecular methods
The Cj1584c amplification primers were designed using the published sequence of the C. jejuni NCTC 11168 [15], and used to amplify the coding sequence of Cj1584c and clone it into protein expression vectors pET-19b and pGEX-6p-2 (Supplementary Table 2).
Table Supplementary 2: PCR primers used in this study.
| Primer | Sequence | Comments |
| 1584pET-F | 5’ ACATATGATGCTTAGATGGTTTGTTTTAC 3’ | NdeI site in bold type |
| 1584pET-R | 5’ ACTCGAGCTATTTACTCCACTCATAAACAT 3’ | XhoI site in bold type |
| 1584pGEX-F | 5’ GAAGAAGAATTCCTGGAATGCTTAGATGGTTTGTT 3’ | EcoRI site in bold type |
| 1584pGEX-R | 5’ TTGTTGCTCGAGCTATTTACTCCACTCATAA 3’ | XhoI site in bold type |
| 1584mut-F | 5’ GTTGGAATTTTGGAC 3’ | [14] |
| 1584mut-R | 5’ CCTTGTAAGCAAAGAC 3’ | [14] |
Recombinant plasmids were transformed into E. coli DH5α host strain, purified and sequenced to confirm the integrity of the sequence. Recombinant plasmids have then been transformed into E. coli BL21 (DE3) to express the His- or GST-tagged proteins. Small scale protein expression and Western blot analysis have been used to confirm the expression of the protein according to [16].
His- and GST-tagged protein purification
The both proteins His and GST tagged proteins were expressed by inducing the cells at OD600 0.4-0.6 with 1mM of Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 hours at 37ºC. His-select nickel affinity gel (Bio-Rad) has been used to purify the His-tagged NikZ protein and the pure protein dialysed against phosphate-buffered saline pH 7.4. GST-tagged NikZ protein has been purified using sepharose 4 fast flow resin (GE Healthcare). The His- and GST-tagged NikZ protein purification have been confirmed by SDS-PAGE gel and Western blot analysis [16].
Amino acids, glycan and small molecules arrays
His-tagged NikZ protein was purified and its ligand binding specificity interrogated using the array analysis. The amino acid, glycan and small molecule arrays were prepared as described previously [17]. His-tagged NikZ was pre-complexed with primary, secondary and tertiary anti-his antibody. The antibodies used were Penta His alexa fluor 555 conjugate. The slides were scanned via proscan array scanner.
Protein-protein interactions
Purified GST-tagged NikZ protein was used to identify protein-protein interaction between the NikZ and the His-tagged periplasmic domain of the Group A transducer like proteins of C. jejuni, Tlps. A 96 well plate had been coated with the GST-tagged NikZ and blocked with blocking buffer 3% (w/v) skim milk powder in TBST (Tris-buffered saline, 0.1% Tween 20) His-tagged Tlp1, Tlp2, Tlp3, Tlp4, Tlp7 and Tlp10 proteins have been added to the wells in presence and absence of nickel for one hour at room temperature. The wells were washed three times with 1x TBS-Tween20. The binding was detected by Anti-His antibody (Cell Signaling Technology).
NikZ-Mutant strain
The NikZ mutant was kindly provided by D. Kelly, Department of Molecular Biology and Biotechnology, University of Sheffield [14]. The mutation was done by cloning the chloramphenicol acetyl transferase (cat) cassette into into ClaI site in cj1584c. The mutation was confirmed PCR.
Growth rate comparison of wild-type and NikZisogenic mutant strains
The wild type and Mutant bacteria strains at OD600, adjusted to 0.05, was inoculated into Muller-Hinton broth, supplemented with 0, 0.1, 0.5 mm, 1 mm and 2 mm nickel and appropriate antibiotics (Vancomycin 10μg/mL and trimethoprim 2.5μg/mL) and grown micro aerobically. The OD had been read every two hours using the spectrophotometer (Eppendorf BioPhotometer Plus) until the culture reached stationary phase. The cell number was confirmed by viable count.
The modified Hard-Agar Plug (t-HAP) assay
The modified Hard-Agar Plug (t-HAP) assay was performed as described previously [18]. Briefly, wild type C. jejuni and the NikZ- isogenic mutant cells numbers were adjusted to OD600 1, 7.5ml of bacterial cells and mixed with 7.5ml 0.6% agar in PBS to final cell number of 1 X 109 cells/ml. Two types of plugs were used in this test, one plug containing 100 mm serine, the second plug containing serine plus nickel chloride at concentrations 0.1, 0.5, 1 and 2 mm. 2,3,5-Triphenyltetrazolium chloride (TTZ) (Sigma-Aldrich) had been added to detect the utilization of serine by the bacteria. The PBS plug had been used as a control. The plates have been incubated micro aerobically for three hours. The plugs were then removed and incubated in Muller Hinton broth for 30 minutes at 42ºC. The cell numbers were determined by viable count.
RESULTS
C. jejuni chemo tactic response to Ni
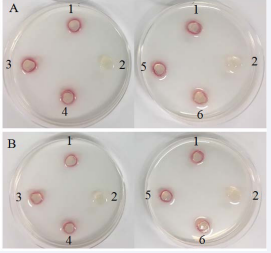
t-HAP chemo tactic assays [18]. were employed to assess chemo responses of the wild type and NikZ- isogenic mutant strains of C. jejuni 11168-O to nickel at 0.1mm, 0.5mm, 1mm and 2mm concentrations. t-Hap assay enables assessment to both chemo attractants and chemorepellent where retardation of migration towards a strong attractant, mixed with a compound being assessed, indicates that the compound, in this case nickel, is a repellent. Chemotactic responses of both wild type and mutant C. jejuni cells demonstrated that Ni acted as a chemorepellent for C. jejuni 11168-O (Figure 1).
Figure 1: t-HAP chemotaxis assay of C. jejuni 11168-O and NikZ isogenic mutant A: T-HAP assay results for C. jejuni 11168-O wild type B: T-HAP assay results for the NikZ- isogenic mutant. (1) 100 mm serine, (2) PBS, (3) 100 mm serine + 0.1 mm nickel, (4) 100 mm serine + 0.5 mm nickel, (5) 100 mm serine + 1 mm nickel, (6) 100 mm serine + 2 mm nicke
The percentage of cell migration inhibition varied from 23.4% to 96.5 % as shown in (Supplementary Table 1).
Table Supplementary 1: Bacterial strains and plasmids.
| Strain/Plasmid | Discerption | Source/reference |
| C. jejuni NCTC 11168-O | Original clinical isolate of C. jejuni11168 | [19] |
| NikZ isogenic mutant | Chloramphenicol cassette has been inserted in the gene nikZ | [14] |
| E. coli DH5α | F– , ø80dlacZΔM15, Δ(lacZYA-argF) U169, deoR, recA1, endA1, hsdR17(rK– , mK+ ), phoA, supE44, λ–, thi-1, gyrA96, relA1 | [20] |
| E. coli BL21 (DE3) | F- ompT hsdSB (rB - mB - ) gal dcm (DE3) | Novagen |
| pGEM-T easy | To do primary cloning of the gene Cj1584c | Promega |
| pET-19b | To express His tagged recombinant NikZ | Novagen |
| pGEX-6P-2 | To express GST tagged recombinant NikZ | GE Healthcare |
The retardation of migration increased with the increase in Ni concentration. The percentages of inhibition in migration of the wild type cells were 23.4%, 51.9%, 84.4% and 88.2% to plugs containing nickel at concentrations of 0.1 mm, 0.5 mm, 1 mm and 2 mm respectively, while the NikZ- isogenic mutant cells showed migration inhibition rates of 48.4%, 55.5% 78.6% and 96.5% to the same nickel concentrations. Statistically there were no significant differences in inhibition of migration between the wild type and NikZ- isogenic mutant cells at the Ni concentration 0.1mm, 0.5 mm and 1 mm while at the concentration 2 mm the NikZ- isogenic mutant appeared to be more sensitive.
Ligand binding and protein-protein interactions involving NikZ protein
Purified His-tagged recombinant NikZ protein had been interrogated to determine if it can bind compounds other than Ni using small molecule array displaying ~400 compounds, including amino acids and glycans (described in [17]). His-tagged NikZ showed no detectable binding to any compound on the arrays. Furthermore, GST-tagged NikZ did not interact with the purified periplasmic sensory domains of the group A Tlp chemoreceptor’s, in presence or absence of nickel, indicating that NikZ protein is unlikely to interact with the chemosensory receptors to induce indirect sensing of nickel.
Effect of Ni on Growth rate of C. jejuni wild type and NikZ- mutant strains
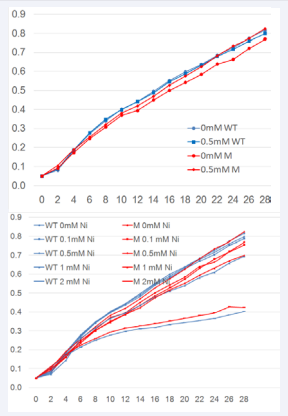
In order to ascertain that the growth rate of NikZ- isogenic mutant strain was not affected by presence of Ni to a greater degree than that of wild type cells, growth rates of both isogenic strains were compared. The presence of nickel decreased the growth rate of the wild type bacteria at all concentrations tested: 0.1 mm, 0.5 mm, 1 mm and 2 mm; it also decreased the growth rate of the NikZ- isogenic mutant at the concentrations 0.1 mm, 1 mm and 2 mm. Interestingly the 0.5 mm nickel increased the NikZ- isogenic mutant’s growth rate to that similar of the wild type strain growth rate in absence of nickel (Figure 2a,b).
Figure 2: Growth rate of C. jejuni 11168-O wild type and NikZ- isogenic mutant. A: The growth rate for C. jejuni 11168-O wild type and NikZ isogenic mutant at nickel concentrations of 0, 0.1 mM, 0.5 mm, 1 mm and 2 mm B: Growth rate C. jejuni 11168-Owild type and NikZ- isogenic mutant at 0 and 0.5 mM nickel concentration
DISCUSSION
This study assessed the role of nickel and NikZ protein in C. jejuni chemotaxis. Although nickel is essential for activation of hydrogenase and urease enzymes in both E. coli and H. pylori, it acts as a chemorepellent at high concentrations [10,11].
The NikZ protein, previously described as nickel binding protein in C.jejuni, did not bind periplasmic sensory domains of the Group A chemoreceptor’s or any of the 400 small molecule ligands on the arrays, suggesting that NikZ is unlikely to play a role in C. jejuni chemotaxis, similar to that of NikA, the Ni-binding protein in E. coli [10].
Similar to other bacteria, nickel was found to be a chemorepellent in equal measure for both, the wild type C. jejuni and NikZ- isogenic mutant strains. The results of t-HAP assay showed significant decrease in C. jejuni cell migration at all nickel concentrations tested (0.1 mm, 0.5 mm, 1 mm and 2 mm) (Table 1). Furthermore, there were no significant differences between the migration inhibition at 0.1, 0.5 and 1 mm nickel concentrations in both wild type and NikZ- , while at the concentration 2 mm of nickel, NikZ- isogenic mutant appeared more sensitive than the wild type. These data are in agreement that NikZ has no role in chemotaxis of C. jejuni, either by direct or indirect sensing as removing this protein did not affect the ability of bacteria to sense nickel, similar to that shown for E. coli [10].
Growth rates for both the wild type and the NikZ- isogenic mutant decreased in presence of nickel at concentrations 0.1 mm, 1 mm and 2 mm while the concentration 0.5 mm boosted the growth rate of the mutant and made it nearly equal to that of wild type, in agreement to that reported previously [14]
CONCLUSION
In conclusion, nickel acts as a chemorepellent for C. jejuni and its effect increases with higher Ni concentration. Furthermore, NikZ- protein has no role in nickel sensing in C. jejuni.
Table Supplementary 1: Bacterial strains and plasmids.
| Strain/Plasmid | Discerption | Source/reference |
| C. jejuni NCTC 11168-O | Original clinical isolate of C. jejuni11168 | [19] |
| NikZ isogenic mutant | Chloramphenicol cassette has been inserted in the gene nikZ | [14] |
| E. coli DH5α | F– , ø80dlacZΔM15, Δ(lacZYA-argF) U169, deoR, recA1, endA1, hsdR17(rK– , mK+ ), phoA, supE44, λ–, thi-1, gyrA96, relA1 | [20] |
| E. coli BL21 (DE3) | F- ompT hsdSB (rB - mB - ) gal dcm (DE3) | Novagen |
| pGEM-T easy | To do primary cloning of the gene Cj1584c | Promega |
| pET-19b | To express His tagged recombinant NikZ | Novagen |
| pGEX-6P-2 | To express GST tagged recombinant NikZ | GE Healthcare |
Table Supplementary 2: PCR primers used in this study.
| Primer | Sequence | Comments |
| 1584pET-F | 5’ ACATATGATGCTTAGATGGTTTGTTTTAC 3’ | NdeI site in bold type |
| 1584pET-R | 5’ ACTCGAGCTATTTACTCCACTCATAAACAT 3’ | XhoI site in bold type |
| 1584pGEX-F | 5’ GAAGAAGAATTCCTGGAATGCTTAGATGGTTTGTT 3’ | EcoRI site in bold type |
| 1584pGEX-R | 5’ TTGTTGCTCGAGCTATTTACTCCACTCATAA 3’ | XhoI site in bold type |
| 1584mut-F | 5’ GTTGGAATTTTGGAC 3’ | [14] |
| 1584mut-R | 5’ CCTTGTAAGCAAAGAC 3’ | [14] |