Molecular Models of Protein Interaction in Photosynthetic Electron Transport
- 1. Department of Biophysics, Lomonosov Moscow State University, Russia
Abstract
The processes of electron transfer in photosynthetic chain at the molecular and subcellular level are simulated. Movements and interactions of individual electron carrier proteins are described by Brownian dynamics, conformational changes accompanying the process of forming a proteinprotein complex are simulated using molecular dynamics methods. Multiparticle Brownian models explicitly reproduce Brownian diffusion of mobile protein carriers and their electrostatic interactions with multi-enzyme complexes both in solution and in the interior of a photosynthetic energy-transforming photosynthetic biomembrane. The analysis of these models reveals the role of diffusion and electrostatic factors in the regulation of electron transport, the influence of geometry of the reaction volume, ionic strength, and pH of the cell medium on the rate of electron transport reactions between electron carrier proteins. The article is based on the results of research carried out at the Department of Biophysics, Faculty of Biology, Lomonosov Moscow State University.
KEYWORDS
- Electron transfer, Photosynthetic chain, Molecular dynamics, Brownian dynamics, Protein-protein complex, Electrostatic interactions, Photosynthetic biomembrane
CITATION
Riznichenko G, Kovalenko IB, Fedorov VA, Khruschev SS, Rubin AB (2024) Molecular Models of Protein Interaction in Photosynthetic Electron Transport. Chem Eng Process Tech 9(2): 1091.
INTRODUCTION
Photosynthesis is a basic process of the existence of life on Earth. This is the only process in living systems where energy is accumulated; all other processes occur with energy consumption. The primary processes of converting absorbed light quanta into the energy of macroergic compounds occur in subcellular systems - the thylakoids of chloroplasts of green plants and algae and the chromatophores of photosynthetic bacteria. Due to the primary processes of photosynthesis, solar energy is converted into the energy of chemical bonds and the oxygen we breathe is released [1-3]. The uniqueness of this process is associated with light-induced electron transfer against the thermodynamic potential along the so-called "photosynthetic electron-transport chain", which results in photosynthetic phosphorylation and reduction of pyridine nucleotides - products necessary for biosynthesis. (Photosynthesis: Molecular Approaches to Solar Energy Conversion. Springer, 2021).
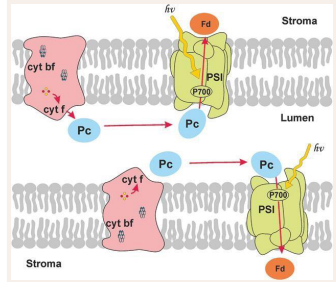
In terms of the possibilities of mathematical modeling, the system of primary photosynthetic processes is a unique object. Modern methods of Cryo-electron microscopy and tomography allow us to obtain images of the photosynthetic membrane with a resolution of several angstroms. The rate constants of individual elementary stages of the primary processes of photosynthesis have been experimentally determined both in solutions and in native systems. Short laser flashes lasting from femtoseconds to picoseconds trigger fast primary processes in photosynthetic membranes [4]. Spectral methods (differential spectroscopy, fluorescence methods, electron paramagnetic resonance method) allow real-time recording of kinetic curves and determination of the rates of rapid changes in the states of individual components of the photosynthetic system. The main participants in the process of photosynthetic electron transport are the multienzyme complexes of photosystem II (PSII), the cytochrome complex, and photosystem I (PSI) embedded into the bilayer membrane and provide directed electron transfer across the photosynthetic membrane [Figure 1]. The mediators between the cytochrome complex and PSI are the molecules of the protein plastocyanin (Pc), diffusing in the lumen. The reduction of NADP molecules, necessary in the carbon fixation cycle, is carried out by the molecules of the small protein ferredoxin (Fd), mobile in the stroma. The kinetic parameters of the interaction of complexes with mobile carriers are determined by both the diffusion of the mobile carrier to the corresponding complex and the probability of the “correct” landing of the mobile carrier on the corresponding site on the donor or acceptor side of the complex. An important role here is played by the size, shape, and electrostatic properties of the carrier, mobile in the corresponding compartment, as well as the geometry of the reaction volume. The rates of electron transfer in these areas depends on the spatial organization of the membrane and the nature of the diffusion of carriers, these rates are the object of regulation by the entire cell.
Electron transfer at transmembrane transfer sites within photosynthetic enzyme-protein complexes (tunneling along the "electron path") and between these complexes (diffusion of mobile carriers in volumes limited by membranes of cellular structures) are provided by various biophysical mechanisms. The characteristic times of electron transfer at individual stages included in the system of primary photosynthesis processes differ by several orders of magnitude from picoseconds to microseconds; the system is multi-scale in time [3,4].
Modeling of processes of different nature in a single system of primary photosynthetic processes requires different mathematical and computer approaches, which, having been "tested" in modeling photosynthetic processes, can be successfully applied to model other metabolic processes.
MULTIPARTICAL BROWNIAN MODELS
Kinetic models based on the mathematical apparatus of ordinary differential equations [5,6], are based on the assumption of a homogeneous distribution of the system components in space. It is assumed that the multienzyme complexes PSI, PSII and cytochrome complexes interact with mobile carriers in accordance with the mass action law. Meanwhile, in the interior of the photosynthetic membrane, the interaction of proteins does not correspond to the concepts of free diffusion and random collisions similar to reactions in solutions. The total number of mobile carriers per grain, as well as the number of low-mobility reaction centers in the membrane, is tens to hundreds of molecules, which is significantly less than the number required to implement the concepts of free collisions and the mass action law. Electron microscopy data indicate a dense arrangement of multienzyme complexes in the membrane, with the complexes protruding a considerable distance into the luminal [3,7,8]. This prevents free diffusion of PQ in the intramembrane space [9], restricts the movement of the mobile carrier Pc molecules in the lumen, which transfers electrons from the cytochrome complex to PSI, and the diffusion of Fd in the stromal space, where Fd molecules participate in electron transfer along the linear pathway and the cyclic pathway around PSI. To simulate the formation of a reaction complex of two interacting proteins in solution in order to predict the structure of the complex and estimate the rate constant of its formation, Brownian dynamics (BD) models are used, based on the mathematical apparatus of Langevin equations, which describe the translational and rotational motion of proteins under the action of random Brownian forces and electrostatic interactions [10-13].
We develop many-body Brownian models to describe the diffusion of mobile protein carriers both in solution and in the interior of the photosynthetic membrane [14-17]. In these models, proteins in quantities of tens to hundreds of molecules per reaction volume can be mobile, like Fd and FNR in the thylakoid stroma, or one of the proteins can be mobile like Pc in the thylakoid lumen, and the second can be part of a relatively immobile multienzyme complex embedded in the membrane-the Cyt f protein is part of the cytochrome b6f complex [Figure 1].
Figure 1: Scheme of electron transport processes in photosynthesis. Two thylakoid membranes and the luminal space between them are shown. Multienzyme complexes of photosystem 1, photosystem 2 and the cytochrome complex are embedded into the membrane. Molecules of the mobile protein plastocyanin (PC) diffuse in the luminal space. Arrows show the electron transfer pathways. The connection with the Calvin cycle is carried out by the protein ferredoxin (Fd) or flavodoxin (Fd). Proteins for whose interactions multiparticle Brownian models have been developed are highlighted in dark
The basics of the method and the results are described in books [5,17], and original articles [14,15-19]. The multiparticle modeling BD method we are developing allows us to use the advantages of the BD method, which takes into account the role of protein shape and electrostatic interactions in electron transport processes, to study the interaction of not individual proteins, but their ensembles in reaction volumes of complex shape. The model provides a clear three-dimensional visual representation of the dynamics of processes in the system on different spatial and temporal scales, the ability to observe the behavior of individual components and obtain average statistical information for the entire ensemble. An example of a scene in the multiparticle Brownian dynamics model is shown in [Figure 2].
Figure 2: Scene in the multiparticle Brownian dynamics model, a section of the photosynthetic membrane where the mobile protein Pc transfers an electron from the cyt f subunit of the cytochrome complex to the donor part of the FCI [18].
Molecules of electron-carrying proteins perform Brownian motion in the medium and simultaneously experience electrostatic interactions with each other and with the charged surface of the photosynthetic membrane. As confirmed by the results of computational experiments [20], the process of electrostatic orientation significantly (by 1-2 orders of magnitude) increases the observed kinetic constant of the total reaction rate compared to if the proteins collided at random places on their surfaces as a result of purely Brownian motion without preliminary electrostatic mutual orientation. The resulting collision complex can transform over time into the final complex or fall apart under the action of the Brownian force. The formation of the final reaction complex is a complex sequence of processes that ensure conformational correspondence between the molecules of the donor and acceptor proteins. In the final complex, tunneling of an electron between the reaction centers of the donor and acceptor proteins becomes possible. To describe conformational movements in such a reaction complex, it is necessary to use molecular dynamics methods, and to simulate the transfer of an electron from the reaction center of the donor molecule to the reaction center of the acceptor molecule inside the complex, it is necessary to use quantum chemistry methods.
The role of electrostatic interactions in the formation of the oxidation-reduction complex of two proteins was studied in the interaction model of the pair of photosynthetic electron transfer proteins, Pc and Cyt f [12,13,17-19]. Pc and Cyt f proteins are oxidation-reduction partners carrying copper and iron atoms in their reaction centers, respectively. In all higher plants and some algae, physiological function of the Pc protein consists of shuttle electron transfer between the f subunit of cytochrome b6-f complex and PSI. Cyt f of the Cyt b6f-complex contains the Pc binding site. An analysis of several thousand structures showed that during the diffusion process two types of encounter complexes of Pc and Cyt f are formed [20]. In the cluster of "productive" configurations the mutual orientation of the Pc and Cyt f molecules corresponds to the orientation in the functionally active complex, determined experimentally [22]. In the complexes of this cluster, Pc atoms have sufficient mobility for further alignment of the active centers. In the cluster of "non- productive" configurations, the mutual orientation of the Pc and Cyt f molecules does not correspond to the orientation in the functionally active complex, and the Pc atoms have very limited mobility.
BROWNIAN/MOLECULAR MODELS OF PHOTOSYNTHETIC PROTEIN INTERACTION
For electron transfer, it is necessary that conformational changes convert the encounter complex into the final complex, where the reaction centers are at a distance close enough for electron tunneling from the donor molecule to the acceptor molecule. Molecular dynamics provide the means for modeling conformational movements of molecules.
Sequential processes of protein diffusion, their mutual orientation and further conformational “adjustment” leading to the formation of the final complex we modeled in two stages. First, with the Brownian dynamics method we repeatedly simulated the processes of proteins approaching each other and their mutual orientation. Cluster analysis of a large ensemble of tens of thousands of trajectories of Brownian motion revealed various energetically favorable metastable states in the process of formation of a complex of proteins of the electroncarrying proteins.
Second, the final configurations of molecules for the most frequently encountered trajectories were used as initial configurations for molecular dynamics. Computer simulation of the complex formation between photosynthetic proteins Pc and Cyt f was carried out for different types of organisms: higher plants, green microalgae and cyanobacteria Cluster analysis revealed metastable conformations, which were compared with the structures obtained experimentally by NMR [17,19].
Brownian and subsequent MD simulations showed that the role of electrostatic interactions and conformational changes in the formation of complexes of Pc and Cyt f proteins is different in organisms with different forms of photosynthetic apparatus. In higher plants and green algae, electrostatic interactions contribute to the positioning of the Pc molecule near Cyt f heme. Further convergence of protein cofactors and the formation of the final complex occur as a result of rotational movements of the Pc molecule around the binding site.
In cyanobacterium Phormidium laminosum, the complex is formed in a “collisional” way without prior orientation of the molecules. In the case of cyanobacterium Nostoc sp., Pc in the course of the Brownian movement approaches Cyt f in the orientation in which the copper atom is facing the cytochrome f due to long-range electrostatic interactions. It should be noted that the rate constants of Cyt f-Pc complex formation estimated in experiments using spectral methods are significantly lower in cyanobacteria than in higher plants [Figure 3].
Figure 3: A1, B1: central structures of “productive” (A1) and “non-productive” (B1) clusters of encounter complexes of plastocyanin and cytochrome f from higher plants with electrostatic energy of more than 8 kT. A2, B2: distance between copper and iron atoms of Pc and Cyt f, obtained from MD calculations that have productive (A2) and non-productive (B2) clusters as the initial structure. A3, B3: structures of the productive (A3) and non-productive (B3) final complexes obtained from MD calculations. Structures are colored according to the value of the B-factor from emerald (0) to ruby (7263 A2 in A1, B1 and 2074 A2 in A3, B3). The thickness of the lines of protein structures is proportional to the value of the B-factor [17]
In the case of green plants and green microalgae, an ensemble of structures with energy of 8 kT formed in the process of diffusional movement is divided into two significantly different clusters. In the first cluster (Figure 3 A1, 61% of the structures) protein molecules have an orientation close to the experimentally obtained functionally active complex with an electrostatic bond between oppositely charged regions of two proteins. In this case, the region of the Pc molecule which forms an electrostatic bond with Cyt f, has low mobility with respect to Cyt f, while the opposite side of the Pc molecule is subject to fluctuations of greater amplitude. This is confirmed by the results of calculation of the Debye-Waller temperature factor (B-factor), shown in Figure 3, A1: emerald color shows areas with the lowest mobility, ruby color means maximum mobility. Thus in the first cluster, the Pc molecule due to rotation under the action of thermal fluctuations, has the opportunity to take a favorable position relative to Cyt f heme, which promotes electron transfer.
Molecular dynamics of the central structure of the second cluster [Figure 3B] gives a rather stable, but inverted orientation relative to the orientation it has in the NMR-obtained complexes. The structures of the second cluster are unproductive metastable states that cannot be easily destroyed by the action of random Brownian force. Their existence may be the reason for the decrease in the rate of electron transfer between Pc and Cyt f at low ionic strengths as defined in [22,23].
More detailed studies, including the study of the matrix of contacts of individual amino acids of interacting proteins and graphs of changes in the number of contacts during the transformation of the preliminary complex into the final one, made it possible to determine the key amino acid residues involved in the formation of the active complex and to reveal the role of electrostatic and hydrophobic interactions underlying this process [19].
CONCLUSION
In the recent years, due to the rapid development of information technologies and the modeling of various systems, the so-called "agent-based" models have spread which seek to derive the properties of whole complex systems from the properties and types of interactions of "agents", the simplest structural elements that make up this system. In molecular dynamics, agents are atoms or their aggregates (coarse-grained modeling) [24-28], while in Brownian modeling the agents are bio macromolecules as solid bodies [12,13].
To study the biophysical mechanisms of interaction of biological macromolecules, it is necessary to simulate the processes at the subcellular and molecular levels. In this case, an important role is also played by the interior in which such interactions take place. To simulate the interaction of assemblies of macromolecules with limited mobility in a heterogeneous interior, we have been developing an approach of multiparticle Brownian modeling [14-15,18]. The space of simulation forms a three-dimensional computer stage, constructed according to the contemporary data about the spatial organization of the photosynthetic membrane. The simulated molecules which move in accordance with the laws of Brownian dynamics, are oriented relative to each other in electrostatic field created by the molecules themselves as well as by the ionic strength of the medium and charges at the boundaries of the reaction volume. The adequacy of the model is proved by comparison of kinetic characteristics of the simulated processes with the observed experimental data.
Using data from the PDB, we can “directly” calculate the electric potential distribution around each of the interacting molecules and evaluate the role of electrostatic interactions in the process of reaction-diffusion (encounter) complex formation in the oxidation-reduction reaction. Molecular dynamics simulation allows us to follow the conformational movements in the coupled donor and acceptor molecules during the process of final complex formation. When approaching other proteins and complexes, the protein orients itself in the electric field created by these proteins and can take a favorable position for subsequent binding. MD modeling allows us to reproduce the Van der Waals and hydrophobic interactions which arise due to changes in the structure of their hydration shell. Consistent application of Brownian and Molecular Dynamics modeling methods allows us to study the details of the physicochemical mechanisms of protein-protein interactions.
AUTHOR CONTRIBUTIONS
Conceptualization, GR and ABR; methodology, GR, SSK and IBK; investigation, VAF, SSK, GR, SSK; writing-original draft preparation, GR. writing - review and editing, all authors; supervision, ABR; project administration, ABR. All authors have read and agreed to the published version of the manuscript.
ACKNOWLEDGEMENTS
The authors are grateful to the Russian Foundation for Basic Research for many years of support for research on photosynthesis. The research was carried out using the equipment of the shared research facilities of HPC computing resources at Lomonosov Moscow State University.
FUNDING
The research was carried out as part of the Scientific Project of the State Order of the Government of the Russian Federation to Lomonosov Moscow State University no. 121032500060-0 with partial support by the Russian Science Foundation (grant no. 22- 11-00009).
CONFLICTS OF INTEREST
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
REFERENCES
- Nelson N, Yocum CF. Structure and function of photosystems I and II. Annu Rev Plant Biol. 2006; 57: 521-565,
- Barber J. The photosystems: structure, function and molecular biology. 2014.
- Shevela D, Bjorn L, Govindjee G. Photosynthesis: solar energy for life,World Scientific. 2019.
- Rubin A.B. Compendium of Biophysics. 2017.
- Rubin AB, Riznichenko GYu (2014) Mathematical biophysics. 2014; 9: 6.
- Stirbet A, Lazar D, Guo Y, Govindjee G. Photosynthesis: basics, history and modelling, Ann Bot. 2020; 126: 511-537.
- Albertsson P. A quantitative model of the domain structure of the photosynthetic membrane, Trends Plant Sci. 2001; 6: 349-354.
- Dekker JP, Boekema EJ. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys Acta. 2005; 1706: 12-39.
- Kirchhoff H, Mukherjee U, Galla H-J. Molecular architecture of the thylakoid membrane: lipid diffusion space for plastoquinone. Biochemistry.2002; 41: 4872-4882.
- Pearson DC, Gross EL. Brownian Dynamics study of the interaction between plastocyanin and cytochrome f. Biophys J.1998; 75: 2698- 2711.
- Haddadian EJ, Gross EL. Brownian dynamics study of cytochrome f interactions with cytochrome c6 and plastocyanin in chlamydomonas reinhardtii plastocyanin and cytochrome c6 mutants. Biophys J. 2005; 88: 2323-2339.
- Gross EL, Rosenberg I. A Brownian dynamics study of the interaction of Phormidium cytochrome f with various cyanobacterial plastocyanines. Biophys J. 2006; 90: 366-380.
- Khruschev SS, Abaturova AM, Fedorov VA, Ustinin DM, Kovalenko IB, Riznichenko GYu, et al. Brownian–dynamics simulations of protein– protein interactions in the photosynthetic electron transport chain. Biophysics. 2015; 60: 212-231.
- Riznichenko GYu, Kovalenko IB, Abaturova AM, Diakonova AN, Ustinin DM, Grachev EA, et al. New direct dynamic models of protein interactions coupled to photosynthetic electron transport reactions. Biophys Rev. 2010; 2: 101-110.
- Kovalenko IB, Abaturova AM, Gromov PA, Ustinin DM, Grachev EA, Riznichenko GYu, et al. Direct simulation of plastocyanin and cytochrome f interactions in solution. Phys Biol. 2006; 3:121-129.
- Kovalenko IB, Diakonova AN, Riznichenko GYu, Rubin AB. Computer simulation of interaction of photosystem 1 with plastocyanin and ferredoxin. BioSystems.2011; 103: 180-187
- Fedorov VA, Kovalenko IB, Khruschev S, Ustinin DM, Antal TK, Riznichenko GYu, et al. Comparative analysis of plastocyanin- cytochrome f complex formation in higher plants, green algae and cyanobacteria. Physiol Plant. 2019; 166: 320-335.
- Kovalenko IB, Knyaseva OS, Antal TK, Ponomarev V, Riznichenko GYu, Rubin AB. Multiparticle Brownian dynamics simulation of experimental kinetics of cytochrome bf oxidation and photosystem 1 reduction by plastocyanin. Physiol Plant. 2017; 161: 88-96.
- Fedorov VA, Khrushchev SS, Kovalenko IB. Analysis of Brownian and molecular dynamics trajectories of to reveal the mechanisms of protein-protein interactions. Computer Research and Modeling. 2023; 15: 723-738.
- Khruschev SS, Abaturova AM, Fedorov VA, Kovalenko IB, Riznichenko GYu, Rubin AB. The identification of intermediate states of the electron transfer proteins plastocyanin and cytochrome f diffusional encounters. Mol. Phys. 2015; 60: 513-521.
- Ubbink M, Ejdebeck M, Karlsson BG, Bendall DS. The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics. Structure. 1998; 6: 323-335.
- Meyer TE, Zhao ZG, Cusanovich MA, Tollin G. Transient kinetics of electron transfer from a variety of c-type cytochromes to plastocyanin. Biochemistry. 1993; 32: 4552-4559.
- Kniazeva OS, Kovalenko IB, Abaturova AM. G Iu Riznichenko, Grachev E A, Rubin A B. Multiparticle computer simulation of diffusion and interaction of plastocyanin with cytochrome f in the electrostatic field of the thylakoid membrane. Biophyzika. 2010; 55: 259-268.
- Karplus M, Petsko GA. Molecular dynamics simulations in biology. Nature. 1990; 348: 631-639.
- Drior RO, Dirks RM, Grossman JP, Xu H, Shaw DE. Biomolecular simulation: a computational microscope for molecular biology. Annu Rev Biophys. 2012; 41: 429-445.
- Childers M, Daggett V. Insights from molecular dynamics simulations for computational protein design. Mol Syst Des Eng. 2017; 2: 9-33.
- Barber J, Ruban AV. Photosynthesis and bioenergetics, World Scientific. 2017.
- Rubin A, Riznichenko GYu. Modeling of the primary processes in a photosynthetic membrane. In: Laisk A, Nedbal L, Govindjee (eds) Photosynthesis in Silico: Understanding Complexity from Molecules to Ecosystems. Advances in Photosynthesis and Respiration. 2009; 29: 151-176.
![Figure 2 Scene in the multiparticle Brownian dynamics model, a section of the photosynthetic membrane where the mobile protein Pc transfers an electron from the cyt f subunit of the cytochrome complex to the donor part of the FCI [18].](https://www.jscimedcentral.com/public/assets/images/uploads/image-1737446203-1.PNG)
![Figure 3 A1, B1: central structures of “productive” (A1) and “non-productive” (B1) clusters of encounter complexes of plastocyanin and cytochrome f from higher plants with electrostatic energy of more than 8 kT. A2, B2: distance between copper and iron atoms of Pc and Cyt f, obtained from MD calculations that have productive (A2) and non-productive (B2) clusters as the initial structure. A3, B3: structures of the productive (A3) and non-productive (B3) final complexes obtained from MD calculations. Structures are colored according to the value of the B-factor from emerald (0) to ruby (7263 A2 in A1, B1 and 2074 A2 in A3, B3). The thickness of the lines of protein structures is proportional to the value of the B-factor [17]](https://www.jscimedcentral.com/public/assets/images/uploads/image-1737446212-1.PNG)








































































































































