Cloning and abiotic stress expression pattern analysis of mulberry MYB gene and its prokaryotic expression in E. coli
- 1. School of Biology and Technology, Jiangsu University of Science and Technology, Sibaidu, Zhenjiang 212018, Jiangsu Province, P. R. China
- 2. Sericultural Research Institute, Guangxi Zhuang Autonomous Region, Nanning,530007, China
- 0. These authors contributed equally to this work
INTRODUCTION
Environmental stress comprising low temperature, high salinity and drought affect plant growth, development, and productivity. To adjust to these adverse environmental stresses, plants have developed complex signaling mechanisms to regulate the expression of stress-related genes that help them withstand and improve stress tolerance either directly or indirectly [1].
Several proteins and genes in the complex signaling networks are regulated by many transcription factors (TFs) families. MYB transcription factor (TFs) exists extensively in eukaryotes. It is one of the abundant and most diverse families of TFs in the plant kingdom. The primary MYB gene (v-myb) was isolated from the avian myeloblastosis virus (AMV), which encodes the MYB domain protein[2-4]. The first plant MYB gene was cloned in maize (Zea mays COLORED1 (C1) gene), which encodes proteins for regulating anthocyanidins [5,6]. Several MYB genes have been identified from plants as an increasing number of plant genomic sequences became available. For instance, in the Arabidopsis thaliana genome, 198 MYB genes have been identified [2,4]; in Brassica rapa, 256 MYB genes have been reported [7].
MYB proteins contain a highly conserved DNA-binding domain (the MYB domain), varying from one to four imperfect amino acid sequence repeats [1,8]. The MYB gene in plants is classified into four distinct groups depending on the number of adjacent repeats present. These include MYB-related (MYBR, containing only one R1- or R2-like repeat), R2R3-MYB (comprising two R2/3R-like repeats), 3R-MYB (involving three R1/R2/R3-like repeats), and rare MYB proteins (4R-MYB, four R1/R2-like repeats; CDC5-like) [2,4]. The genome-wide analysis of the MYB TFs reveals that all four groups of MYB proteins are found in plants. According to research, the largest and the most extensively studied group of the MYB genes in plants is the R2R3- MYB having over 100 members, which have been extensively amplified many years ago[9]. For instance, in O. sativa, A. thaliana, and Glycine max 113, 126, and 244 genes have been encoding for R2R3-MYB proteins, respectively [4,5,10]. Although the role of several MYB genes has been categorized in many plants, the function of 4R-MYB proteins remains uncertain. MYBR proteins play an active role during cellular morphogenesis, organ morphogenesis, secondary metabolism, phosphate starvation, regulation of the circadian gene, and chloroplast development [2,11]. The R2R3-MYB proteins have been shown to play crucial roles in several biological processes, including development, responses to various biotic and abiotic stress, and metabolism (in phenylpropanoid metabolism) [12].
Mulberry (Morus spp.) is a perennial tree or shrub, economically grown for sericulture production as the only food for the domesticated silkworm and other uses, including edible fruits, useful timber, medicinal applications, and ornamental value [13]. The Mulberry plant is susceptible to biotic and abiotic conditions, and its growth and productivity are adversely affected by abiotic and biotic stresses [5,13]. Higher plants always adjust to the environmental stresses by activating various molecular machinery involved in stress perception, signal transduction, and expression of specific stress-related genes. Since MYB TFs play an essential role in plant-specific processes such as primary and secondary metabolism, developmental function, and response to biotic and abiotic stress [5], they were studied in many plant species. However, there are very few reports on the functional characterization of MYB TFs in the mulberry plant. The study by [5] revealed the MYB3R1 transcription factor in Morus. notabilis (encoded by MnYB3R1) binds to the MnPPO1 promoter region and implies that MnYB3R1 and PPO are both involved in the abscisic acid-responsive stress response pathway. Therefore, it is imperative to investigate the MYB TFs in the mulberry plant response to abiotic conditions.
In this study, the MYB TFs gene was cloned from cDNA of young fresh leaves of Mulberry (YU-711) based on Mulberry expressed sequence tags (ESTs) using homologous cloning technology and exploiting reverse transcriptase-polymerase chain reaction (RT-PCR). Changes in the transcription level of the MYB gene under drought, cold, and salt stresses were detected by qRT-PCR at various time points. The findings will lay the foundation for understanding the complicated signal transduction mechanism of MYB superfamily underlying stress responses and the basis for molecular improvement on mulberry resistance through gene regulation technology and the breeding of resistant varieties.
MATERIALS AND METHODS
Plant growth and stress treatment
The Mulberry (Morus multicaulis) variety YU-711 was grown under a controlled environment at the National Mulberry Gene Bank of the Sericultural Research Institute, Chinese Academy of Agricultural Sciences, Zhenjiang. R. China. The grafted seedlings were planted and later transferred into 35cm diameter pots. Each pot comprising a seedling was grown in an incubator with a 24 h photoperiod (12 hours per day and 12 hours of darkness) at a constant temperature of 25°C. After the shoots have grown to 20cm in height, the seedlings were exposed to various stress treatments such as drought, cold, and salts [14]. Mulberry seedlings were subjected to water deficit for 14 days to simulate drought stress, while the control group was watered daily. Leaves were sampled at (0d 2d, 4d, 6d, 8d, and 10d) after water deficit based on preliminary results got on this species [14]. Mulberry leaves sampled were quick-frozen in liquid nitrogen and stored at -80? for total RNA extraction. For the salt treatment, the mulberry seedlings were watered with 0.3 mol/L salt solution (300 mM NaCl) for 0d, 12h, 1d, 2d, 3d, 4d, and 5d while the control group was treated with sterile water. Mulberry leaves were sampled, frozen in liquid nitrogen, and stored at -80? for total RNA extraction. For the low temperature treatment, Mulberry seedlings were grown under 4°C for 1d, 2d, 3d, 4d, 5d, and 6d, however, the control group was grown under standard temperature at 25°C; leaves were sampled and immediately frozen in liquid nitrogen and stored at -80 degrees for total RNA extraction.
RNA isolation and cloning of the cDNA
Total RNA was extracted from the leaf (net weight approx. 100 mg) of the mulberry seedlings under various treatments, as mentioned above. RNAiso Plus reagent (Takara, China) was employed. Following the manufacturers’ protocol, treated with Deoxyribonuclease I and then resuspended in 0.1% (v/v) DEPCtreated water and kept at -80°C. The total RNA was measured with 1.0% (w/v) agarose gel electrophoresis; the total RNA’s quality was determined with an ultraviolet spectrophotometer. Using the RNase H-Reverse Transcriptase M-MLV kit (Takara, China) protocol, the first-strand cDNA was synthesized from the total RNA. The first-strand complementary DNA as the template for PCR in gene cloning. Primer 5.0 software was used to design the MYB gene primers according to the EST as a forward primer (MYB-F-5?ATTGTCCCTCACTATCCGTA-3?), reverse primer (MYBR-GGTTGGGTCACCTCTTTTGC 3?) with the inference function from mulberry cDNA library and the homology sequences from the NCBI (http://www.ncbi.nlm.nih.gov/) databases using the BLAST program. The amplification program used for the PCR is 94°C pre-denaturation for 6 minutes; 94°C for 25 seconds, 62°C for 30 seconds, 72°C for 1 minute, 25 cycles; 72°C extension for 4 minutes. The products were separated on 1.0% agarose gels and sequenced by Sangon Biotech (Shanghai, China). Thus, a cDNA sequence of the MYB was derived according to the fragment assembly.
Bioinformatics analysis of the cDNA sequence of the MMYB gene
The NCBI online Search Tool NCBI (http://www.ncbi.nlm. nih.gov) was used to conducts a homology comparison of target genes and the sequence analysis of target genes for identity and similarity. The sequence encoding for the amino acids was analyzed using the ORF finder program at NCBI. Using Uniprot (http://www.uniprot.org/), PlantTFDB database (http:// planttfdb.cbi.pku.edu.cn/) (http://gsds.cbi.pku.edu.cn/index. Php), (http://www.ebi.ac.uk/intact/), PID (http://www.plapid. org), the protein structural and functional domains of the MYB gene were analyzed. NCBI/blastp and NCBI CDD (http://www. ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) were used to verify and conserve domain of the protein was determined. NCBI/ SMART blast was used for the systematic evolution analysis of the gene. ProtParam tool of ExPASy (http://web.expasy.org/ protparam/) was used to study the characteristics of the encoding protein by predicting the molecular weight and theoretical isoelectric point (PI) of the protein.
Expression analysis of MYB by qRT-PCR
The qRT-PCR method was employed to analyze the expression level of the MMYB gene from leaves sampled between different stress treatments. Total RNAs were isolated from leaves of various treatment conditions using RNAiso Plus reagent (Takara, China) according to the manufacture’s protocols and then quantified by computing the absorbance at 260 nm [14]. 1µg of the various RNA samples was used as a template for the synthesis of the cDNA, as mentioned earlier. The final product was then diluted 12-fold, and 3µl of the cDNA was used as the template for the qRT-PCR analysis.
The SYBR Green RT-PCR assay was made in a Light Cycler® 96 Real-Time PCR System (Roche, USA) by three-step real-time qPCR according to the FastStart Universal SYBR Green Master Mix kit (Roche, USA) protocol. The thermal cycling parameters for qRT-PCR was 95°C for the 30s followed by 40 cycles of 95°C for 5s, 55°C for 10 s, and 62°C for 30s using the primer MYB-F (5?-ATTGTCCCTCACTATCCGTA-3’) and MYB-R (5’- GGTTGGGTC ACCTCTTTTGC, 3?).
The melting curves were constructed during amplification. Actin3 (HQ163775.1); PP2A (XM_01091140.1) and Ubiquitin-like protein 5 (XM_010091673.1) as the reference gene for drought, salt, and cold stress, respectively [15]. Actin3; F(5’-GAGGGCCGTGTTCCCCAGCATCGTC-3’), R(5’- TCTTTTTGATTGAGCCTCATCCCCT-3’), PP2A; F (5’-GAA GTT CAT GGTGCATCGGC-3’), R (5’-TCA AAG GCA AAGCAAACCCG-3’), Ubiquitin-like protein 5; F (5’-AGA CCT GAA GAAGCT CGT GG-3’), R (5’-TAG TAG AGC TCGAGG CCC ATC-3’) as a standardization control for each starting quantity cDNA template. All reactions were examined in three biological repeats. The relative expression differences of mRNAs in leaves of different treatments were calculated by the 2−ΔΔCt method [16].
Abiotic stress assays
The expression level of the MMYB mRNA under abiotic stresses (drought, salt, and low temperature) in Mulberry was measured by qRT-PCR. All the stress treatments, for the same time interval, were carried out three times. The two pairs of qRTPCR primers [14] (stated above) were used for the quantitative real-time PCR to elucidate the relative expression values. The reaction program was not different from those mentioned above.
Construction of expression vectors for recombinant MMYB gene
Total RNA was isolated from fresh samples using RNAiso Plus reagent (Takara, China) following manufacturer protocol. The first-strand cDNA was synthesized using Prime Script™ RT Reagent Kit (Takara) according to the instructions by the manufacturer. Forward and reverse primers of the ORF of the MYB gene were designed and synthesized to contain sequences of Hind III at the beginning of the forward primer MYB F (5’-CCAAGCTT ATTGTCCCTCACTATCCGTA-3?) and EcoR1 at the end of the reverse primer MYB R (5 ?- CGGAATTC GGTTGGGTCACCTCTTTTGC 3?). The ORF of the MYB gene with the Hind III and EcoR1 as restriction enzyme sites was amplified from the cDNA by PCR. The amplification conditions for the PCR are as follows; 94°C for 6 mins; 30 cycles, 94°C for 25s, 62°C for 30s, 72°C for 1 min, and 72°C for 4 mins. The PCR products were separated on 1.0% agarose gels, recovered, ligated with pUC19-T vector (Takara, China), and then transformed into E. coli DH5α competence cell. The positive clones were selected after culturing with amp+ and verified by PCR before sequencing. The restriction enzyme Hind III and EcoR1 were used in digesting the MMYB fragment and the expression vector pET-30a (+). The digested were then ligated together with T4 DNA Ligase. The resultant recombinant plasmid pET-30a (+)-MYB was identified by PCR, enzyme cleavage, and sequencing to confirm the correct orientation of the MYB gene.
Prokaryotic expression of MMYB in E. coli
The extracted recombinant plasmid (pET-30a (+) with MMYB) was transformed into E. coli Rosetta-gami B. The positive clone of E. coli Rosetta-gami B -pET-30a (+)-MMYB was cultured in LuriaBertani (LB) broth containing 50 µg/ml kanamycin overnight (about 12 h) at 37°C by shaking at 200 rpm. A Fresh LB medium (3 ml) containing kanamycin was mixed with 100µl aliquots of the overnight culture and incubated at 37°C on a 200rpm shaker until an OD600= 0.5-0.8 was derived. The culture was mixed with IPTG to a final concentration of 0.5 mM to induce the recombinant protein expression and incubated on a shaker at 160 rpm for 8h at 37°C. The bacterial cells were attained by centrifuging at 8,000 rpm for 5 min, at 4°C. The cell pellet was washed by 1× phosphate buffer saline (PBS) (pH7.4) three times and then subjected to sonication at 500 W for 6 times with 5 s working and 5 s free on ice. It was centrifuged at a speed of 10,000rpm for 5 min at 4°C. The supernatant was preserved directly at -20°C, and the precipitate kept at -20°C after resuspended by 1 × PBS.
SDS-PAGE and Western Blot detection
According to the Tricine-SDS-PAGE protocol, SDS-PAGE was used to analyzing the recombinant protein [17] as described by [14]. The precipitate together with the protein samples was mixed with 2 × SDS-PAGE loading buffer (0.05 M Tris. HCl, pH 6.8, 10% glycerol, 2% SDS, 0.01% bromo phenol blue, 0.1 M β-ME) and boiled for 15 min at 95°C. the samples were loaded on a 4% stacking gel-12% separating gel, and the gel running was got according to the manufacturer’s instructions (BIO-RAD) after centrifugation at 12,000rpm for 3 min. Electrophoresis was performed by 80V-150V voltage, and then the protein samples were detected.
With the western blotting analysis, the non-stained SDSPAGE gels were transferred onto the PVDF membrane by electro blotting using Mini Trans-Blots Electrophoretic Transfer Cell System (BIO-RAD). The membrane was blocked in 7.5 ml TBST (0.01 M Tris-HCl, pH7.5 + 0.15MNaCl+0.1% Tween-20) solution containing 0.6 g bovine serum albumin for 1h at 37°C by shaking. After being washed with TBST three times at 8min intervals, the membrane was incubated with mouse anti-6His Tag primary antibody at 1:1000 dilution in TBST for one hour in darkness at 37°C with shaking. The membrane was then washed three times with TBST and incubated with alkaline phosphatase (AP) conjugated goat anti-mouse antibody at 1:2000 dilution in TBST for 1h in darkness at 37°C with shaking. Immunoreactivity was detected using the ECL chemiluminescence imager detector.
RESULTS AND ANALYSIS
Cloning and analysis of the mulberry MYB gene
Information on the EST with the inference function, from the mulberry cDNA library and sequences of the conserved regions of known MYB genes from other plant species, the Mulberry’s MYB gene’s cDNA was cloned by RT-PCR and then sequenced. The obtained cDNA of the MYB gene has an open reading of 882bp. The ORF encodes for a protein of 293 amino acid residuals with a predicted molecular weight of 41.7 kDa and a theoretical isoelectric point (PI) of 7.61. The mulberry MYB gene belongs to the SANT superfamily (Figure S2. (g)). With NCBI blast (http:// blast.ncbi.nlm.nih.gov/Blast.cgi#), the corresponding mulberry gene (Morus 006161.t1.e2) was determined. The expected length of the SANT’s MYB (XP_010109645.1) gene is 1648bp. Homologous analysis using blast (http://blast.ncbi.nlm.nih. gov/Blast.cgi#)) reveal that the mulberry MYB gene has a 97% identity to the SANT MYB gene.
The SMART prediction of the MYB protein sequence of Mulberry showed that the MMYB protein has two SANT domains at the N-terminus and C-terminus, respectively. The SANT domain occurs at amino acid on 61st arginine (R) site to the 116th aspartic acid (D) site and the other SANT at the of lysine (K) with at position 177th to arginine (R) at 230 sites (Supplementary S2) of the protein sequence.
Homologous evolution analysis of mulberry MYB gene
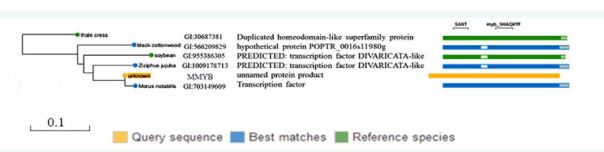
To further analyze the mulberry MYB transcription factor’s sequence characteristics and functions, SMART blast analysis of the mulberry MYB gene was carried out. We selected the three best homologous matching genes with MYB and two best matching genes of the species that have been studied through SMART blast. Using the SMART blast, first, we used BLAST for a non-redundant (nr) protein database to optimize search query for the MYB gene sequence. BLASTP was then used to identify homologous sequences that are like MYB. The COBALT multiple sequence alignment program was used to perform multiple sequence alignments based on conserved protein domains between MMYB and the five selected sequences and produce the best alignment among all sequences. The phylogenetic relationship, as shown in Figure 3. The Phylogenetic relationship was constructed to determine the relationship between MMYB and other biological species. SMART blast provides the alignment of the MMYB gene sequence with five homologous matching sequences, namely: Populus trichocarpa, Jinsixiaozao, soybean, Arabidopsis thaliana, and Chuan mulberry.
SMART blast shows that the MMYB gene has a conserved domain. The Chuan Sang, Populus trichocarpa, and Jinsixiaozao were the three best matches between MMYB and nonredundant protein sequences. The homologous genes have conserved SANT and myb_SHAQKYF domains. The MMYB gene (unknown protein) has a close relationship with Chuan Sang (Morus notabilis). The corresponding conserved domain transcription factor is the Chuan Sang. The other species that have a relationship is Arabidopsis, whose conserved domain is duplicated homeodomain-like superfamily protein (At2G38090), Populus trichocarpa, Jinsixiaozao, and soybean, are predicted and uncharacterized proteins.
Expression profiles of MYB gene under salt, drought, and low temperature stress treatments
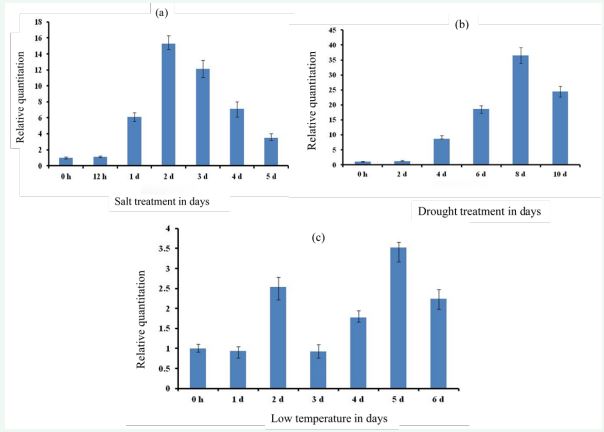
To elucidate whether abiotic stress induces the MMYB gene expression, the mRNA transcript level of the MMYB gene under different abiotic stress treatments, including salt, drought, and cold, was measured at different time points by qRT-PCR analysis. The findings showing some variations in the results are presented in Figures 1 (a-c).
Figure 1 The mRNA expression levels of Mulberry MYB gene under abiotic stress treatment in different periods. The horizontal axis represents the time of abiotic tresses, and the vertical axis represents the relative expression level of MYB gene. The results are MYB mean ± SD of the triplicates of three biological replicates. (a) Expression profile of the MYB gene under salt stress; the expression levels of MYB were normalized to the level of control reference (PP2A; XM_01091140.1). (b) Expression profile of the MYB gene under drought stress; the expression levels of MYB were normalized to the level of control reference (Actin 3, HQ163775.1). (c) Expression profile of the MYB gene under cold stress; the expression levels of MYB were normalized to the level of control reference (ubiquitinlike protein 5, XM_010091673.1
Figure 1a illustrates the effect of salt stress treatment on mRNA expression of the MMYB gene. The MMYB gene expression level at 12h after salt stress was low with a minimum of about 1-fold same as the level of the control. The expression level then increased sharply on day one with a level of 6.1 times that of the control and then continues to increase on day two, reaching an expression maximum of 15.8 times that of the control. After day two, the expression level sharply declined to 12 times than the control on the third day (3d). The expression level drastically reduced from day three and again to the fifth day, with an expression level of 4 times than the control. The expression level and pattern clearly show that the MYB gene expression is induced in the mulberry plant’s early stage under salt stress. The evidence shows that MMYB genes are involved in response to salt stress. These findings conform to the work of [18], which reports that the MYB gene has a positive regulator of salt stress tolerance response in transformed Arabidopsis plants. Again, this result is consistent with [19] on the regulatory role of the MYB gene in response to salt stress in transgenic rice.
Figure 1b illustrates the effect of drought stress treatment on the mRNA expression of the MMYB gene. The MMYB expression level at day two 2d was at a minimum of about 1.5 times as the control from the figure. The expression level rapidly increased from day four (4d), recording 10 times compared to the control at 0 h. Again, the expression level rose to about 17 times on 6 d and reached a maximum expression level on the eight days (8 d) with a sharp increment, recording 37 times the level of the control. There was a drastic decline from 8 d to 10 d with an expression level 25 times compared to the control, which indicate that the MYB gene is induced and is positively regulated in response to drought stress plant growth and development [20].
Figure 1c illustrates the effect of low temperature stress on mRNA expression of the MMYB gene. There was transcript expression fluctuation. The MMYB gene expression level reduced to about 0.9 times the level of control over the first day after the tress treatment (1 d). There was a rapid increase in the expression level of 2.7 times the control from the first day to the second day. Then again, the expression attains a sharp decline of 0.9 times the control on the third day.
A steady increase in transcript level was reached on the fourth day with 1.7 times that of the control. A sharp increase was observed on day 5 recorded the maximum transcript expression of 3.7 times that of the control and sharply to its maximum. A sharp decline occurred from the fifth day to the sixth day, with an expression level of 2.4 times that of the control. The MMYB gene was highly induced and regulated, explaining that the MMYB gene is highly involved in response to cold stress. These results are also consistent with the findings reported by[19] and [1].
Expression of MMYB gene in E. coli and western blotting detection of recombinant protein
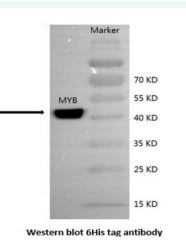
The recombinant plasmid pET-30a (+) - MMYB was transformed into an E. coli for the expression of MMYB after induction by IPTG for 3h; the recombinant protein of the MMYB was highly and effectively expressed in E. coli Rosetta gami. The protein (MMYB) has a molecular weight of 41.7 kDa and a (PI) of 7.61, consistent with the predicted molecular weight and PI using bioinformatics and detected by SDS-PAGE analysis. The recombinant protein induced by the IPTG was transferred into the PVDF membrane after SDS-PAGE separation and was detected by western blot using mouse anti-6His Tag primary antibody. As depicts in Figure 3, the protein band was clearly defined by western blot and estimated to be 41.7 kDa of molecular weight.
DISCUSSION
Mulberry (Morus spp.), a perennial tree or shrub, is an economically vibrant crop grown not only for sericulture production as the only food plant for the domesticated silkworm but also for a variety of other uses, such as the production of edible fruits, useful timber, medicinal applications, and ornamental value. Adverse environmental factors such as biotic and abiotic stresses affect Mulberry plants during growth and developmental stages [13]. Therefore, it is vital to identify the primary stress tolerance mechanisms in Mulberry and developing strategies for genetic improvement of abiotic stress resistance. We isolated and cloned the MYB gene from the cDNA of Mulberry variety YU711. The MYB transcription factor(TF) belongs to the SANT superfamily, an extensive protein family in the plant kingdom known to perform a crucial role in response to abiotic stress such as cold, salinity, and drought in plants [1]. For the first time, the MMYB gene that cold for the SANT domain was got from the Mulberry variety YU-711. The sequence analysis reveals that the obtained sequence had an Open Reading Frame (ORF) of 882bp, encoding 293 amino acids. The Molecular weight and a theoretical isoelectric point were predicted to be 41.7kDa and 7.61, respectively. Bioinformatics analysis shows that the mulberry MYB protein domain is highly conserved and contains SANT and myb-like DNA-binding domain, SHAQKYF class, belonging to the SANT superfamily (Supplementary S1).
MYB-like DNA-binding domain with SHAQKYF class is restricted to or common in plants protein. It contains a response regulator domain which appears associated with the myb-like DNA-binding domain distinguished in part by well-conserved motif SH[AL]QKY[RF] at the C-terminal end of the motif [21]. MYB proteins are described based on their highly conserved DNA-binding domains. Depending on the number of imperfect repeats of the SANT (for SWI3, ADA2, N-CoR, and TFIIIB) domain usually of 50–53 amino acids in MYB DNA-binding domains, plant MYB proteins can be subdivided mainly into three subfamilies consisting, MYB-related ( single SANT domain), the R2R3-MYB (two SANT domains) and R1R2R3-MYB (three SANT domains) [1,8]. Gene expression in leaf veins is usually regulated by the change in environments and physiological, metabolic signals of this tissue during leaf development and growth[22]. This may explain that MMYB gene might function in vascular tissues in leaves during stress response. Studies have reported on the role of MYB TFs in plant abiotic stress. In AtMYBC1, a 1R-MYB protein was reported to be a repressor of freezing tolerance in a CBF independent pathway in Arabidopsis [1,23]. MYBS3 has been reported in rice to play an essential role in cold stress tolerance [24]. This conforms to our studies, as the MMYB gene expression under cold stress increased (figure 3c) compared to the control showing that the MMYB gene is actively involved in cold stress response.
Plants under high-salt environments generate primary stress signals and secondary signals, including hyperosmotic signals, which leads to the accumulation of ABA, which activates the ABA signaling pathway and triggers the expression of downstream stress response genes, and hence leading to enhanced plant salt tolerance [18,25]. In our work, qRT-PCR analysis reveals that the MMYB was highly expressed under salt stress treatment (figure 3a). This indicates that MMYB may be associated with a salt stress response. In soybeans, GsMYB15 has been reported to act as a regulator to regulate the expression of other genes or interact with other TFs as TF complex to affect downstream gene expression, positively regulating salt stress tolerance [18]. Plant MYB proteins have been reported as major key player in Arabidopsis. For instance, AtMYB2 and AtMYB15 are reported to play a key role in ABA-dependent drought stress responses [20]. Again Over-expression of TaMYB30-B and TaMYB19-B is reported to improve drought stress tolerance in transgenic Arabidopsis [26]. Further studies reveal that AtMYB96 act as a molecular link that mediates ABA-auxin crosstalk in drought stress response and lateral root growth, offering an adaptive strategy under drought [27].
A phylogenetic tree was built using the COBALT multiple sequence alignment program with the MMYB and other five species’ amino acid sequences. These species include Populus trichocarpa, Jinsixiaozao, soybean, Arabidopsis thaliana, and Chuan Mulberry (Figure 2).
Figure 2 The phylogenetic tree based on the amino acid sequence of the MYB from Mulberry (Morus multicaulis) and other homolog sequences from 5 different species. The evolutionary tree was established in the COBALT multiple sequence alignment program using the maximum likelihood method. Green color shows a match from a reference species, the blue color shows a match from a non-redundant protein database, and yellow color indicates a target gene
The evolutionary analysis showed that MMYB gene is homologous to Chuan Sang, Jinsi Xiaozao, and Populus trichocarpa. However, the relationship with Chuan Sang (Morus notabilis) is the closest.
E.coli (BL21(DE3)) is most extensively used in molecular biology and for the production of recombinant proteins because of its high efficiency of expressing any gene of interest that is under the control of T7 promoter [28]. Under the activity of T7 RNA polymerase, when an inducer such as IPTG or rhamnose is added to the culture, T7 RNA polymerase is expressed and transcribes the gene of interest, followed by a translation of the desired protein by using endogenous protein translation machinery [28]. On this basis, MMYB gene could bind to BL21 T7 promoter and with the T7 RNA polymerase, and addition of IPTG at a concentration of 0.5 mM, the desired protein was induced and expressed. As it was prdicted by ExPASy (http://web. expasy.org/protparam), we successfully expressed the protein fused with the tagged protein in the E.coli Rosetta B strain. To produce a recombinant protein of MMYB with a molecular weight of 41.7kDa and separated by SDS-PAGE and confirmed with western-blotting detection (Figure 3).
Figure 3 Western blot detection of the MaMYB fusion protein expressed in E. coli. Marker: protein molecular weight standard; MYB: total MMYB protein of E. coli Rosetta-gami B harboring pET30a-MMYB induced by IPTG. Arrow indicates the band of the MMYB protein.
In conclusion, we obtained valuable evidence on the MMYB gene; we reveal with qRTPC that MMYB gene was expressed in response to various abiotic stresses such as salt, drought and low temperature as studied in this work. We successfully expressed the MMYB gene protein in E. coli cell and with SDS-PAGE and Western blot, we deduce the molecular weight of the protein to be 41.7kDa as predicted by bioinformatics, ExPASy (http:// web.expasy.org/protparam). Elucidating the MYB protein and regulation that is possible in Mulberry will provide the basis for the function for predicting the contribution of MYB proteins to the biology of Mulberry plant during breeding. In future studies, different MYB gene in Mulberry will be investigated.
ACKNOWLEDGMENTS
T This work was supported by the earmarked fund for CARS18, Key R&D Program of Guangxi(2022AB19054), Zhenjiang Science and Technology support project (GJ2021015), the Crop Germplasm Resources Protection Project of the Agriculture Ministry (111721301354052026), and National Infrastructure for Crop Germplasm Resources (NICGR-43).
Conflict of interest
The authors declare that they have no conflict of interest.
REFERENCES
- Yong, Y., Y. Zhang, and Y. Lyu, A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences, 2019. 20(13): p. 3195.
- He, C., J.A. Teixeira da Silva, H. Wang, C. Si, M. Zhang, X. Zhang, M. Li, et al., Mining MYB transcription factors from the genomes of orchids (Phalaenopsis and Dendrobium) and characterization of an orchid R2R3-MYB gene involved in water-soluble polysaccharide biosynthesis. Sci Rep, 2019. 9(1): p. 13818.
- Klempnauer, K.-H., T.J. Gonda, and J.M. Bishop, Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell, 1982. 31(2): p. 453- 463.
- Li, Y., K. Lin-Wang, Z. Liu, A.C. Allan, S. Qin, J. Zhang, and Y. Liu, Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.). Int J Biol Macromol, 2020. 148: p. 817-832.
- Liu, D., S. Meng, Z. Xiang, G. Yang, and N. He, An R1R2R3 MYB Transcription Factor, MnMYB3R1, Regulates the Polyphenol Oxidase Gene in Mulberry (Morus notabilis). Int J Mol Sci, 2019. 20(10).
- Paz-Ares, J., D. Ghosal, U. Wienand, P. Peterson, and H. Saedler, The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. The EMBO Journal, 1987. 6(12): p. 3553- 3558.
- Wang, Z., J. Tang, R. Hu, P. Wu, X.L. Hou, X.M. Song, and A.S. Xiong, Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genomics, 2015. 16(1): p. 17.
- Dubos, C., R. Stracke, E. Grotewold, B. Weisshaar, C. Martin, and L. Lepiniec, MYB transcription factors in Arabidopsis. Trends Plant Sci, 2010. 15(10): p. 573-81.
- Feller, A., K. Machemer, E.L. Braun, and E. Grotewold, Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J, 2011. 66(1): p. 94-116.
- Katiyar, A., S. Smita, S.K. Lenka, R. Rajwanshi, V. Chinnusamy, and K.C. Bansal, Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics, 2012. 13: p. 544.
- Ambawat, S., P. Sharma, N.R. Yadav, and R.C. Yadav, MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants, 2013. 19(3): p. 307-21.
- Liu, J., A. Osbourn, and P. Ma, MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol Plant, 2015. 8(5): p. 689-708.
- Chen, D., A. Justice Afriyie, R. Li, K. Dominic, L. Li, R. Li, and W. Zhao, Molecular cloning of potassium transporter gene, MaHAK5 of mulberry (Morus alba L.) and gene expression and biochemistry analysis under potassium stress. The Journal of Horticultural Science and Biotechnology, 2019. 94(1): p. 130-136.
- Li, Y., J. Zhang, F. Hu, A. Adolf, M. Ackah, A.A. Justice, Q. Lin, et al., Cloning and abiotic stress expression analysis of galactose-binding lectin (GBL) gene from mulberry and its prokaryotic expression in E. coli. The Journal of Horticultural Science and Biotechnology, 2020: p. 1-10.
- Shukla, P., R.A. Reddy, K.M. Ponnuvel, G.K. Rohela, A.A. Shabnam, M.K. Ghosh, and R.K. Mishra, Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Mulberry (Morus alba L.) under different abiotic stresses. Mol Biol Rep, 2019. 46(2): p. 1809-1817.
- Schmittgen, T.D. and K.J. Livak, Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc, 2008. 3(6): p. 1101-8.
- Schägger, H., Tricine–sds-page. Nature protocols, 2006. 1(1): p. 16.
- Shen, X.-J., Y.-Y. Wang, Y.-X. Zhang, W. Guo, Y.-Q. Jiao, and X.-A. Zhou, Overexpression of the Wild Soybean R2R3-MYB Transcription Factor GsMYB15 Enhances Resistance to Salt Stress and Helicoverpa Armigera in Transgenic Arabidopsis. International Journal of Molecular Sciences, 2018. 19(12): p. 3958.
- Tang, Y., X. Bao, Y. Zhi, Q. Wu, Y. Guo, X. Yin, L. Zeng, et al., Overexpression of a MYB Family Gene, OsMYB6, Increases Drought and Salinity Stress Tolerance in Transgenic Rice. Frontiers in Plant Science, 2019. 10.
- Wang, N., W. Zhang, M. Qin, S. Li, M. Qiao, Z. Liu, and F. Xiang, Drought Tolerance Conferred in Soybean (Glycine max. L) by GmMYB84, a Novel R2R3-MYB Transcription Factor. Plant Cell Physiol, 2017. 58(10): p. 1764-1776.
- Lu, S., J. Wang, F. Chitsaz, M.K. Derbyshire, R.C. Geer, N.R. Gonzales, M. Gwadz, et al., CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res, 2020. 48(D1): p. D265-d268.
- Wu, X., R. Huang, Z. Liu, and G. Zhang, Functional characterization of cis-elements conferring vascular vein expression of At4g34880 amidase family protein gene in Arabidopsis. PLoS One, 2013. 8(7): p. e67562.
- Zhai, H., X. Bai, Y. Zhu, Y. Li, H. Cai, W. Ji, Z. Ji, et al., A single-repeat R3-MYB transcription factor MYBC1 negatively regulates freezing tolerance in Arabidopsis. Biochem Biophys Res Commun, 2010. 394(4): p. 1018-23.
- Su, C.-F., Y.-C. Wang, T.-H. Hsieh, C.-A. Lu, T.-H. Tseng, and S.-M. Yu, A Novel MYBS3-Dependent Pathway Confers Cold Tolerance in Rice. 2010. 153(1): p. 145-158.
- Leng, P., B. Yuan, and Y. Guo, The role of abscisic acid in fruit ripening and responses to abiotic stress. J Exp Bot, 2014. 65(16): p. 4577-88.
- Zhang, L., G. Liu, G. Zhao, C. Xia, J. Jia, X. Liu, and X. Kong, Characterization of a Wheat R2R3-MYB Transcription Factor Gene, TaMYB19, Involved in Enhanced Abiotic Stresses in Arabidopsis. Plant and Cell Physiology, 2014. 55(10): p. 1802-1812.
- Baldoni, E., A. Genga, and E. Cominelli, Plant MYB Transcription Factors: Their Role in Drought Response Mechanisms. 2015. 16(7): p. 15811-15851.
- Yamazaki, D., T. Itoh, H. Miki, and T. Takenawa, srGAP1 regulates lamellipodial dynamics and cell migratory behavior by modulating Rac1 activity. 2013. 24(21): p. 3393-3405.