RNAi Regulation Profile Comparison in SH-Sy5y and U87 Cells: Response upon ? Endorphin
- 1. College of Medicine & Medical School, Yonsei University, Seoul, Korea
- 2. Seoul National Univeristy Hospital Biomedical Research Insitutue, Seoul, Korea
- 3. Korean Language Major & Pre Medicine Department of Arts and Sciences, University of Washington, USA.
- 4. Department of Mathematics, Yonsei Univeristy, Seoul, Korea
Abstract
Glioblastoma (GBM) is an important tumor that has prevalence in childhood over adults, whereas many other types of tumor have high incidence after adolescence. It is a rare disease in terms of epidemiology of tumor incidence by age groups. The key role of synthetic endorphin in its RNA modification level is investigated in glioblastoma and neuroblastoma cell model by micro RNA array profiling. Our findings indicate that endorphins have highly skewed transcriptome processing profile in neuroblastoma as compared to glioblastoma. Also, cell viability decreased in Glioblastoma upon beta endorphin treatment. It provides a transcriptional model for endorphin neurotransmitter effects with two human brain derived cells.
Keywords
- RNAi target
- Glioblastoma
- Endorphin
- Neuroblastoma
- Profile dependent indicator
Citation
Kim S, Im W, Shulipa O, Kim B (2017) RNAi Regulation Profile Comparison in SH-Sy5y and U87 Cells: Response upon β Endorphin. Int J Rare Dis Orphan Drugs 2(2): 1008
INTRODUCTION
Endorphins belong to a group of neurotransmitters known as opioids, which are peptide like signaling molecules that bind to their receptor in the brain. The receptor functions as a G-coupled protein complex with k, u, and m subunits, and belongs to a group of heptahelical model proteins that are expressed in both the central and peripheral nervous systems. Other opioids such as enkephalins and dynorphins also bind opioid receptors [1,2].
Most of the opioid-endorphin categorical neurotransmitters are reported with sensory function and pain [3]. In case of rats, stress has shown to decrease the malignant tumor resistance mediated by the release of opioid peptides [4]. In a study on stress and its effect on malignant tumor development, it was shown that stress decreases the opioid mediated tumor resistance in rats. Previous research done by Tug McGraw Research Center reported that subjects with brain cancer tend to have high levels of psychological stress [5], whereas, Université de la Méditerranée (France) pilot research data reports that this ‘stress’ induce the higher rates of brain tumors. In the use of morphines, glioblastoma proliferation is impaired [6].
Cerebral cancer treatments, such as brain surgery and total-ectomy of partial brain tissues, has shown high disability rates in post-treatment patients. Treatment with radiotherapy or chemotherapy carries a high risk of compromising the patients’ immune system, resulting in a heightened sensitivity to nosocomial pathogens.
GBM is one of the common malignant tumors with 5 year survival rate less than 5% [7,8,9]. Malignant glioblastoma is common after age 50 with WHO standard Grade IV, which has high resistance to chemotherapy [10]. U87MGcell line is derived from these tissues have studied thoroughly to find the anticancer drug’s therapeutic effects [11,12,13,14].
Compared to the established cancer treatments, surgery and radiotherapy, pre-existing material control was intended to minimize the side effects. Especially, we applied materials that are present in the human nervous system, such as neurotransmitters. In case of endorphin, its effect on reduction of stress and increase of immunity is well known [15,16], and this endorphin requires opioid receptors [17,18].
In this study, we postulated that there will be miRNA level changes which may be underlying within the cancer inhibitory mechanisms. Endorphins naturally exist as neurotransmitters, which is naturally degraded in polypeptide form. Therefore, our study focuses on profiling miRNA interactions with endorphin as a key growth controlling factor in U87 glioblastoma and SH-Sy5y neuroblastoma cells.
MATERIALS AND METHODS
Cell culture and induction of neural process formation
SH-SY5Y, a human neuroblastoma and U87 glioblastoma cell line (ATCC, Manassas,VA, USA), were used in this study. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 100 U/mL of penicillin, 0.1 mg/mL of streptomycin (Sigma-Aldrich, St Louis, MO, USA), and L-glutamine (2 mM) in 75-cm2 tissue culture flasks (Falcon, Becton Dickinson, Lincoln Park, NY, USA) at 37o C in 5% CO2 . SH-SY5Y and U87 cells were incubated with synthetic beta endorphins at 0.1μM, 1μM and 10μM concentration. This was analyzed with WST-1 viability assay (Sigma-Aldrich). The source of the cells was Homo sapiens ‘either sex’ (male and female).
RNA isolation
Total RNA was extracted from SH-SY5Y and U87 cells treated with 1μM of endorphin for 48 hours by using Trizol (Invitrogen) using the manufacturer’s protocol. Addition of chloroform followed by centrifugation separates aqueous and organic phase. RNA was recovered by precipitation with alcohol. Starting with 1,000 ng/μL RNA as template, cDNA was synthesized by using 50 ng/μL random hexamers, 0.1 M dNTPmix, and DEPC-water at 65o C 5 min. 10 RT buffer, 25 mM MgCl, 0.1 M DTT, RNase OUT, and Superscript III RT (all from Invitrogen) were added in the final volume of 20 μL, and the reaction was incubated at 25o C for 10 min, 50o C for 50 min and 85o C for 5 min.
MicroRNA array and Transcriptome profiling.
Total RNA was isolated from cells using Trizol (Invitrogen, Carlsbad, CA, USA). The RNA purity and concentration were measurement using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Tech. Rockland, DE, USA) and a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Expression profiles for miRNA were examined using an Agilent Mouse miRNA Microarray 8×15 K kit, which detects 567 mouse miRNAs, according to the manufacturer's instructions (Agilent Technologies). Two RNA samples were hybridized for each group. Equal amounts of total RNA from each sample were used. Scanning and analysis were performed using an Agilent hardware platform, and the data were normalized and analyzed using GeneSpring GX, version 7.3.1 (Agilent Technologies) according to the manufacturer's instructions. Measurements less than 0.01 were set as 0.01. The total amounts of miRNAs in each sample were determined by summing the levels of each miRNA.
Statistical methods.
Student’s t-test was used to evaluate the inhibitory effect of endorphin and its significant role in dose dependent manner. ANOVA and Chi-square test were also applied to table sets in RNAi gene expression profile.
RESULTS
Viability effects of synthetic endorphin in U87 cell line
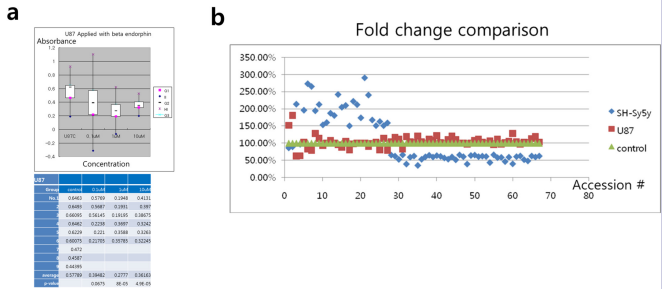
The endorphins reduced the viability of U87 GM cells. Compared to control, there was a significant decrease in proliferation after the treatment of endorphin (Fig. 1a).
Figure 1: The proliferation inhibitory effect in treatment of beta endorphin in U87 cells (a) Dose dependent effects while the dose increase between 0.1μM and 1μM (P = 8E-05), but did not show differences between 1μM and 10μM. (b) Fold change comparison in micro RNA targets in U87 and SH-Sy5y cells. 67 accession # were screened and roughly changes over 1.5 fold change are enlisted. The fold change remained with a high alteration in SH-Sy5y cells, but did not show a great difference in U87, compared to control. No significant change was observed in other targets. Both cells had adverse effects in the targets (accession # 1 ~ 67).
As the dose rose to 0.1μM, 1μM and 10μM, the p-value turned as 0.0675, 8E-05 and 4.9E-05 respective to the control group. It showed dose dependent effects while the dose was increased from 0.1μM to 1μM, but did not show significant differences between 1μM and 10μM.
Profile comparison between U87 and SH-Sy5y transcripts
Micro RNA array analysis between the effects of endorphin in U87 and SH-Sy5y cells were adverse. Out of human gene transcripts, 67 micro RNA targets showed change more than 1.5-fold in U87 or SH-Sy5y cells (Fig. 1b). In those 67 targets, 11 targets were up-regulated and 7 targets were down regulated more than 2-fold in SH-Sy5y solely (Table 1 and Table 2).
Table 1: The top 5 highest conserved match throughput in micro RNA targets with higher than 2 fold down regulation, uniquely in SH-Sy5y cells. The changes solely represent effect on SH-Sy5y cells as an inhibitory manners, in miR-708, miR-454, miR-375, miR-532-5p,miR 301b, miR-106b and miR-1248. The targets for miR 301b and miR-454 overlapped according to the TargetScan(Co.) Analysis.
Table 2. The top 5 highest conserved match throughput in micro RNA targets with higher than 2 fold change, uniquely in SH-Sy5y cells. SH-Sy5y cell target micro RNA with over 2 fold up-regulation is enlisted. Zinc Finger protein expression was regulated, in miR-595 and miR-1260. Zinc Finger domain is a key transcription factor for neurotransmitters, which is postulated to alter expression in the behalf of beta endorphin treatment.
|
|
Discovered possible targets for Glioblastoma specific regulations
The hsa-miR-574-3p only showed different and opposing fold change between SH-Sy5y cells and U87 cells. It was up regulated about 2.318 fold in SH-Sy5y cells and 0.625 down regulated in U87 cells. In other words, the micro RNA down regulates the target genes in SH-Sy5y cells and up-regulates in U87 cells (Table 3). The general target genes for hsa-miR-574-3p (3'ACACCCACACACGUACUCGCAC 5’) are retinoid X receptor alpha, Kruppel-like factor 12, cullin 2 (CUL2), DAB2 interacting protein (DAB2IP), NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4-like 2 and FOS-like antigen 2 (Table 3a). In case of cullin 2, nearly 70% of the naturally-occurring cancer-disposing mutations in VHL abrogate elongin binding, suggesting that binding to elonginCUL2 complexes contributes to the ability of VHL to suppress tumor growth in vivo. It is suggested that CUL2 is a candidate tumor suppressor gene, as has been proposed for CUL1. DAB2IP is a Ras GTPase-activating protein (GAP) that acts as a tumor suppressor. The DAB2IP gene is inactivated by methylation in prostate and breast cancers. Expression of DAB2IP was lower in prostate cancer cell lines than in normal prostate epithelial cell lines. They showed that the P1 promoter of DAB2IP was active in normal cells, but inactive in the cancer cell lines (Table 3a).
Table 3: Candidates for differential regulation in between two cell lines. (a) Screened target genes for hsa-miR-574-3p. For Kruppel-like factor 12 (KLF12), the conserved sites were 8-oligomer, but 7-oligomer targets were conserved in retinoid X receptor alpha (RXRA), cullin 2(CUL2), DAB2 interacting protein (DAB2IP), NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4-like 2, and FOS-like antigen 2. (b) In case of miR-498, it was down regulated for SH-Sy5y and upregulated in U87 MG. Its target genes coded non-human origin transcripts, which show variation in mitochondrial genes and chromosome 9 related transcripts
DISCUSSION
Treatment of cells with synthetic beta neurotransmitter has shown to inhibit cell proliferation as determined by cell count and WST-1 assay. In the course of human micro RNA target analysis, it has been discovered that U87 cells might have possible unique properties that are coded by the has-miR-574-3p gene sequence. The changes solely represent effect on SH-Sy5y cells as some inhibitory manners, in miR-708, miR-454, miR-375, miR-532-5p, miR 301b, miR-106b and miR-1248. The targets for miR-301b and miR-454 overlapped (Table 1). Zinc Finger protein expression was regulated, in miR-595 and miR-1260, while Zinc Finger domain is a key transcription factor for neurotransmitters (Table 2).
Opioids are known to reduce pain and it is used in some cases to reduce long-term pain that is exerted by cancer. Also, it play multiple roles in central and peripheral nervous system. Opioids influence immunity, respiration, behavior and intestine movement. However, other than its influence in pain, it is reported to regulate cell growth. Therefore, it should be carefully controlled when performing cancer therapy [1]. Also, it activates the vascular endothelial growth factor (VEGF) receptor, which worsens the status of cancer [19,20]. Thus, we postulated that internal opioids such as endorphin might affect cancer cell growth. As a result, in initial concentration of 0.1 μM, the effects were indistinguishable, but when concentration was increased in a log scale, the proliferation was decreased. Previous studies showed that some opioids such as morphine facilitated the growth of glioblastoma T98G in concentrations of 20 and 40 μM but decreased its proliferation on treatment with opioid biphalin. Biphalin is known to have 1000-fold pain reducing effect compared to morphines.
The dose-dependent response is contingent on interactions between ligands and ligand receptors. In low concentrations, these opioids act as an agonist, whereas in higher concentrations they function as antagonist. Higher concentrations alter the receptors configuration causing it to react as an antagonist. Therefore, according to our study, endorphins may be considered as antagonists to their receptors if the concentration is increased, diminishing their effect [25, 26].
In line with signal transductions that are derived from cell membrane proteins, there are several possible mechanisms for cell growth inhibition [26]. For example, although it has been reported that opioids might be able to regulate cell growth or apoptosis [1,21,22,23]. PARP1 inhibition is a possible therapeutic target in GBM [27]. However, in case of T98G glioblastoma cell membrane, opioid receptors are expressed and the expressions did not change regardless of opioid treatment. Thus, it is early to generalize that cell proliferation and apoptotic signaling might be a phenomenon related to down-regulation or desensitization by receptor internalization [24]. Several genes, such as CUL2 and DAB2IP genes are studied as markers in prostate and breast cancers, but should be verified to show a conceptual model in glioblastoma and neuroblastomas. An implication of Ras dependent signaling pathway and related tumor suppression is another issue, its limitations might also be related to synthetic endorphin binding in its opioid receptors.
In conclusion, our new finding supports that transcriptional difference underlies in the effects of endorphin, especially in neuronal origin cells. Unique mechanisms are found in certain micro RNA. Our profiles will supplement a post-transcriptional genetic base for endocrine materials, which are poorly studied for drug resistance, conditions such as placebo response, neuroprotectivity and endorphin neurotransmitter effects of human brain derived cells.
ACKNOWLEDGEMENT
We thank Dr. Jae-joon Bahn for reviewing cell line samples.
DISCLOSURE
The authors declare no competing financial interests.
REFERENCES
25. Sarkar DK, Zhang C. Beta-endorphin neuron regulates stress response and innate immunity to prevent breast cancer growth and progression. Vitam Horm. 2013; 93: 263-276. (doi: 10.1016/B978-0- 12-416673-8.00011-3)
26. Sarkar DK, Murugan S, Zhang C, Boyadjieva N. Regulation of cancer progression by β-endorphin neuron. Cancer Res. 2012 Feb 15; 72(4): 836-840. (doi:10.1158/0008-5472.CAN-11-3292.)
27. Murnyak B, Kouhsari MC, Hershkovitch R, Kalman B, Marko-Varga G, Klekner A, Hortobagyi T. PARP1 expression and its correlation with survival is tumour molecular subtype dependent in glioblastoma. Oncotarget. 2017; 8(28): 46348-46362. (doi: 10.18632/ oncotarget.18013)