Peptide Profiling of IgG and their Interaction with Receptor Outer Membrane Porin of Klebsiella pneumoniae
- 1. Department of Biotechnology, University of Mysore, India
Abstract
Immunoglobulins make up the largest portion of the humoral immunity, of which Immunoglobulin G (IgG) is the main class of antibodies present in ruminant colostrum. Proteolytic enzymes have been successfully used to generate peptides for reagent and therapeutic use. IgG purified from buffalo (Bubalus bubalis) colostrum was proteolytically digested by pepsin and pancreatin to generate peptide fragments and analyzed by nanoLC-MS/MS (Liquid chromatography - mass spectrometry). We identified 25 peptide sequence matches to heavy and light chains in the complimentary determining region of IgG. The small peptides generated were used to examine their interaction on Klebsiella pneumoniae outer membrane protein (OMP), a drug target site. IgG peptides were found to interact with porin emaluating standard antibiotic ampicillin suggesting their probable interaction to the drug target. A more detailed investigation would be interesting to establish newer functionalities to peptides of IgG which can have profound implications in developing therapeutic formulations.
Keywords
Buffalo colostrum; Immunoglobulin G; nanoLC-MS/MS; Peptides; Porin.
CITATION
Aparna HS, Mamatha Bhanu LS (2017) Peptide Profiling of IgG and their Interaction with Receptor Outer Membrane Porin of Klebsiella pneumoniae. JSM Biochem Mol Biol 4(2): 1026.
ABBREVIATIONS
FC: Fragment Crystallizable Region; FAB: Antigen Binding Fragment; F(ab’)2: Two Antigen Binding Fragment; Fd: Heavy Chain; IgG: Immunoglobulin G; VH: Heavy Chain; VL: Light Chain; ESBL: Extended-Spectrum Beta-Lactamases; CDRs: Complemen tarity-Determining Regions, HR-LC-MS: High Resolution-Liquid Chromatography Mass Spectrometry; OMP: Outer Membrane Porin; Gscore: Glide Score
INTRODUCTION
Immunoglobulins (Igs) are glycoproteins that form an important part of the immune system. Ruminant colostrum is characterized by its very high level of immunoglobulin G (IgG) [1]. In colostrum, Igs are the principal agents that protect the gut mucosa against pathogenic microorganisms, and confer passive immunity to the ruminant neonate until its own immune system is developed [2]. Oral administration of immunoglobulin preparations from human serum as well as bovine colostrum and serum have been evidenced to be safe as well as effective in human clinical trials in treating gastrointestinal tract infections [3] and other microbial infections [4]. Hence, colostrum and milk are categorized as medicinal nutrition [5]. IgGs are large proteins of approximately 150 kDa, made of two identical heavy chains (50 kDa) and two identical light chains (25 kDa) [6]. Antibodies recognize foreign molecules (antigens) through epitopebinding sites in the variable domains of the antigen binding fragment (Fab) and alert immune cells to putative threats through interaction sites in the constant domain of the tail region [7]. Because of the hinge region IgG molecules is flexible and solvent-exposed, Fc (fragment crystallizable), Fab (antigen binding fragment), and F(ab’)2 (two antigen binding fragment) fragments can be readily generated by enzymatic cleavage under native condition [8]. IgG molecules composed of only antigen binding portions can be advantageous and have potential for immunotherapeutic applications in cancer treatment, infection clearance, and targeted drug delivery [9]. The complementarity determining regions (CDRs) that reside in the variable regions of light chains and heavy chain (Fd) are responsible for the antigen binding specificity and consequently, have an effect on its potency [10]. The smaller size enables in better tissue penetration thus facilitating better antigen recognition in immunochemistry and less steric hindrance leading to more sensitive antigen detection. The enzyme pepsin cleaves the Fc portion of an immunoglobulin into small sub fragments leaving a F(ab’)2 fragment with two antigen binding sites connected by disulfide bonds [11]. Colostrum IgG, a neonatal food component, survive gasterointestinal digestion was performed by pepsin and pancreatin, the resulted peptide fractions analysed by HR-LC/MS (High performance liquid chromatography mass spectrometry).
The outer membrane of gram-negative bacteria is the first line of defense against toxic compounds and provides a variety of functions including passive and active transport, host pathogen recognition, signal transduction, and catalysis. Due to their exposed epitopes on the cell surface and highly conserved immunogenicity [12,13], Omps could be used as candidates to develop vaccines for combating bacterial infections [14,15]. As a most important member of Omps, the outer membrane protein A (OmpA) is a class of highly conserved proteins among the enterobacteriaceae family [12]. OmpA is confirmed as a multifunctional protein that plays an important role in bacterial physiology and pathogenesis. It can function as an adhesin and invasin, participate in biofilm formation, act as both an immune target and evasin, and serve as a receptor for several bacteriophages [13]. OmpA (outer membrane protein A) is one of the best characterized OM proteins and highly conserved among the Enterobacteriaceae family. K. pneumoniae produces three major proteins, OmpK35, OmpK36, and OmpA, in its outer membrane [14]. Based on the sequence similarities of ompK35 and ompK36 genes, OmpK35 is a homolog of OmpF [15], and OmpK36 [16] is a homolog of OmpC [17]. Clinically, most of the (Extended-spectrum beta-lactamases) ESBL-producing K. pneumoniae strains express only OmpK36. This barrier comprises a lipid bilayer that is impermeable to hydrophobic antibiotics and other compounds [18]. Thus by evaluating interactions of the ligand/s with the receptor with energy-based score or scoring function for each pose, provide clues of the potential bioactive peptide ligands of IgG peptides across OMP. In this context, IgG characterized from buffalo colostrum [19] possessing growth inhibitory potential on K. pneumonia [20] was digested with the enzymes pepsin and pancreatin and the peptides were analysed by nanoLC-MS/MS. Based on the in vitro results, the probable interaction of IgG peptides was studied in silico by targeting outer membrane porin (OMP) of K. pneumoniae.
MATERIALS AND METHODS
In vitro gastrointestinal digestion of IgG
In vitro gastrointestinal digestion was carried out in triplicates as described earlier [21]. A 3.5 % (w/v) IgG sample in 0.1 M KCl HCl (pH 2) buffer was incubated with pepsin (4% w/w) for 4 h at 37°C and the reaction was terminated by keeping in boiling water for 10 min. The suspension was neutralized with addition of 2 N (Normality) NaOH to pH 7 and digested with 4% (w/v) pancreatin at 37°C for 4 h. The enzyme was inactivated by boiling for 10 min and centrifuged (10,000xg, 30 min) and desalted.
HR-LC/MS analysis
The desalinized peptide mixture was fractionated on Agilent 6550 iFunnel QTOF mass spectrometer (Agilent Technologies) coupled to an Agilent G1376A Infinity Capillary Pump and G2226A Infinity Nanoflow Pump LC system (Agilent). The desalted permeate was loaded on to a Protein ID-Chip (G4240-62030 High Performance Chip, 360 nl enrichment column, 150 mm x 75Âμm) via an Infinity Auto sampler (Low Flow Hip Sampler, Agilent Technologies) in buffer A (0.1% formic acid in MilliQ water) at a flow rate of 1 µl/min of Capillary Pump and 0.3 µl/min of nano Pump. Peptides were eluted into the mass spectrometer through a linear gradient with initial conditions of 3% buffer B (90% Acetonitrile, water, 0.1% formic acid) increasing up to 98% buffer B over 20 min. Peptides were introduced to the mass analyzer from the LC through Agilent 1260 Chip-cube operating in ESI-positive (Jet Stream Ionization) ion mode; precursor mass within a range of 300-3200 m/z were selected for MS/MS with a threshold absorbance count of above 2000 and ramped collision energy was limited to charge 2, 3 and more than 3 . The MS scan rate (acquisition rate) was 6 spectra/sec and acquisition time of 166.6 ms. MS/MS spectra were collected with an isolation width at medium (~4 amu) resolution and ramped collision energy for different charge states. MS/MS spectra were scanned from 0-3200 m/z with acquisition rate of 3 spectra/sec and acquisition time of 333.3 ms. Precursor/parent ions were excluded for 0.15 min following selection for MS/MS.
Molecular docking
The peptides obtained from IgG after in vitro gastrointestinal simulation digestion were used for docking analysis targeting outer membrane porin of K. pneumoniae (www.rcsb.org; PDB ID: 1OSM) using Schrodinger software. The protein was prepared for molecular docking by protein preparation wizard for the addition of missing atoms, hydrogens, assigning bond orders, setting proper ionization states of residues and capping the termini. The receptors were then refined with H-bond assignment (water orientations, at neutral pH) and energy was minimized with OPLS 2005 force field. The receptor grid specified was with reference to the residues (Phe 189, Thr 229, Tyr 231, Phe 257; PDBSUM software). The receptor grid scaling of Vander Waals radii was set at 0.8 and partial charge cut off value of 0.15. The ligands (FIFPP, IKSFRI, IVKPGASV, FDVWGTGT) along with standard antibiotic ampicillin (PDB ID: 4GCP) were prepared in ligprep with the following parameters, force field: OPLS2005, ionization at target pH: 7.0 ± 2.0, tautomers and stereo isomers (generate all combinations of specified chiralities and determine chiralities from 3D structure) with at most 6 ligands to be generated. Finally, the ligands were docked by XP (Extra Precision) method with flexibility (nitrogen inversions and ring conformations). The results were exported with XP descriptors information by adding Epik State penalties to docking score.
RESULTS AND DISCUSSION
HR-LC-MS analysis
The purified buffalo colostrum IgG was digested by a combi nation of proteases, pepsin and prancreatin. The resultant pep tides obtained after HR-LC/MS analysis was searched against non redundant NCBI database. Search results revealed the identity of 25 peptide sequences matching to IgG heavy and light chains of mouse, human and bovine species. The identified protonated mo lecular ions matching to IgG light chain were m/z - 1165.5583+, m/z-1576.8006+, m/z-1818.8915+, m/z-2345.0489+ and m/z 3454.6617+ corresponded to LSVSSVTAEDSA, LIYSTNQWP SPAVT, LSPGERATLSCRASHSF, LPGTAAISELQSEDEGDYYCAL and LSVATGEKVTIRCITNTDIDDDMNWYQQKP sequences (Table 1).
|
Table 1: Light chain matched peptide sequences of colostrum IgG treated with pepsin and pancreatin. |
|||||
|
Sl. No. |
M/Z |
Sequence |
Accession number |
Protein |
Organism |
|
1 |
1165.5583 |
LSVSSVTAEDSA |
CCM09771.1 |
Immunoglobulin kappa chain variable region |
Mouse |
|
2 |
1576.8006 |
LIYSTNQWPSPAVT |
BAN63134.1 |
IgG L chain |
Human |
|
3 |
1818.8915 |
LSPGERATLSCRASHSF |
S16832 |
Ig kappa chain V region |
Bovine |
|
4 |
2345.0489 |
LPGTAAISELQSEDEGDYYCAL |
CAC94285.1 |
Immunoglobulin lambda chain variable region |
Human |
|
5 |
3454.6617 |
LSVATGEKVTIRCITNTDIDDDMNWYQQKP |
AAD39779.1 |
Immunoglobulin kappa light chain variable region |
Mouse |
In mammals there are two types of Ig light chains, lambda (λ) and kappa (κ) but only one type of light chain is present in any antibody. Among these peptides, LSVSSVTAEDSA and LSVAT GEKVTIRCITNTDIDDDMNWYQQKP matched to κ chain from mouse, LIYSTNQWPSPAVT and LPGTAAISELQSEDEGDYYCAL matched to λ chain from human while, LSPGERATLSCRASHSF was found similar to bovine κ light chain from variable domain.
Each Ig heavy chain has an N-terminal variable (V) region containing the antigen-binding site and a C-terminal constant (C) region, encoded by an individual C region gene, that determines the isotype of the antibody and provides effectors or signaling functions [21].
The molecular ions m/z - 882.39991+, m/z - 935.495211+, and m/z - 1625.88051+ were matched to sequences – FDVWGTGT, FI ISRDNA and WVRGVPEKGLEWVA homologues to V-D-J-region. VDJ gene segments encode the variable region of the heavy chain. V (D) J recombination is the primary mechanism for diversifica tion of the human antibody repertoire which allow rapid humoral immune responses to a wide range of pathogenic challenges [24]. The peptide sequences FIFPP (m/z - 619.33696+), IKSRFI (m/z - 763.48319+), IVKPGASV (m/z - 769.46976+), LLKPSETLSL (m/z - 1100.65685 2+), LKSRLSIMKD (m/z - 1190.69325+), WIRQAPG KGL (m/z - 1124.64543+), IVQPGGMKLSCA (m/z - 1202.61511+), WVRQVPEKGL (m/z - 1210.68221+), IRQPPGKGLEWIM (m/z - 1523.82821+), LSLTCTTSGFSLS (m/z - 1316.64095+), LVKQSQTLSLTCT (m/z - 1421.76755+), LESLTCSVSGGSIS (m/z - 1339.64167+), LRDTSVTAAL (m/z - 1046.58475+), VKQRPG GGLGWIF (m/z - 1414.79606+), ISDYQMNWIRQTGKGLEWLYM (m/z - 2632.26932+), IRQPPGKGLEWIGCVSDSGSTSYYPT (m/z - 2697.2984+), and FTFSDYYMAWVRQVPEKGLEWVANINYDGSST (m/z - 3859.6617+) matched to the Ig heavy chain variable re gion. Most of the sequence identity was from human Ig and a very few matches were from mouse and bovine species (Table 2).
|
Table 2: Heavy Chain matched peptide sequences of colostrum IgG treated with pepsin and pancreatin |
||||
|
Sl. No. |
M/Z |
Sequence |
Accession number |
Protein |
|
1 |
619.336962 |
FIFPP |
AAP205491 |
Immunoglobulin G heavy chain |
|
2 |
763.48319 |
IKSRFI |
AAD24790.1 |
Immunoglobulin heavy chain variable region |
|
3 |
769.46976 |
IVKPGASV |
CAC29289.1 |
Immunoglobulin heavy chain |
|
4 |
882.39991 |
FDVWGTGT |
AEC22872.1 |
Immunoglobulin heavy chain V(D)J region |
|
5 |
935.49521 |
FIISRDNA |
AAA37969.1 |
Immunoglobulin heavy chain V-D-region |
|
6 |
1046.58475 |
LRDTSVTAAL |
AAR32583.1 |
Immunoglobulin heavy chain |
|
7 |
1100.65685 |
LLKPSETLSL |
JH0428 |
Ig gamma chain V region (VH4-T14) |
|
8 |
1124.64543 |
WIRQAPGKGL |
AIT38804.1 |
Immunoglobulin G heavy chain variable region |
|
9 |
1190.69322 |
LKSRLSIMKD |
AAA98646.1 |
Immunoglobulin heavy chain variable region |
|
10 |
1202.61511 |
IVQPGGMKLSCA |
AGA92777.1 |
Immunoglobulin heavy chain |
|
11 |
1210.68221 |
WVRQVPEKGL |
AAK19265.1 |
Immunoglobulin heavy chain variable region |
|
12 |
1316.64095 |
LSLTCTTSGFSLS |
ABF48111.1 |
Immunoglobulin gamma heavy chain |
|
13 |
1339.64167 |
LESLTCSVSGGSIS |
CEF91268.1 |
Immunoglobulin heavy chain |
|
14 |
1414.79606 |
VKQRPGGGLGWIF |
AAG39133.1 |
Immunoglobulin heavy chain |
|
15 |
1421.76755 |
LVKQSQTLSLTCT |
AAC48521.1 |
Immunoglobulin heavy chain variable region |
|
16 |
1523.82821 |
IRQPPGKGLEWIM |
AET35388.1 |
Immunoglobulin G heavy chain variable region |
|
17 |
1625.88051 |
WVRGVPEKGLEWVA |
AAB48326.1 |
Ig heavy chain V-D-J-region |
|
18 |
2632.26936 |
ISDYQMNWIRQTGKGLEWLYM |
AAQ05424.1 |
Ig heavy chain variable region, VH3 family |
|
19 |
2697.2984 |
IRQPPGKGLEWIGCVSDSGSTSYYPT |
CAD82966.1 |
Ig heavy chain |
|
20 |
3859.6617 |
FTFSDYYMAWVRQVPEKGLEWVANINYDGSST |
ADM44572.1 |
Immunoglobulin G1 heavy chain V- D-region |
However no data is available on the buffalo IgG either from serum or milk in the existing database. Hence these sequences can be uploaded to the protein database. This method can be exploited for generating antibody fragments with potential use as drugs, so-called single-domain antibodies [22]. In analogy to our ap proach, a more economical and efficient production of mouse monoclonal IgG2a F(ab’)2 fragment was developed by Stowers [9] for immunotherapeutic applications.
The use of antibodies in immunoassays such as ELISA or western blotting is critical for antigen detection. However, sometimes it is beneficial to use a portion of the molecule instead of the whole antibody itself. By selectively cleaving an antibody with proteases and using reducing agents, it is possible to engineer fragments with discrete characteristics. Following reduction of disulfide bonds, three antibody domains (LC, Fd, and Fc/2) were characterized by LC-MS, capillary isoelectric focusing and glycan mapping [23]. It is also possible with genetic engineering techniques to express small fragments directly. In some cases it may be desirable to remove the Fc domain, which may mediate unwanted binding events, leaving a bivalent Fab’2 fragment and Fab’ is important for better tissue penetration (derived from Fab’2) or Fab may be preferred. Antibody fragments are commonly used as the starting point for drug molecules as a result of having lower immunogenicity than intact antibodies. Therefore sequential digestion by pepsin and pancreatin was found to enhance the effectiveness and economic feasibility of passive oral immunoprophylaxis with bovine milk Igs [24].
Molecular docking
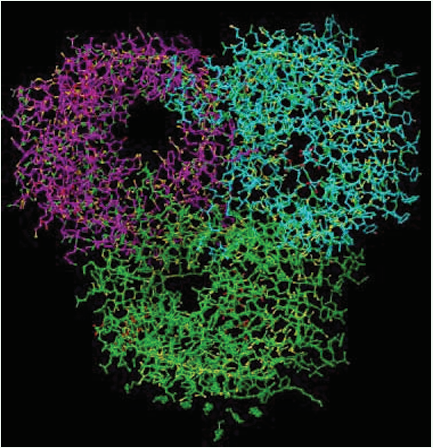
The small peptides (5-8 residues) obtained after in vitro gastrointestinal digestion of IgG were used to examine their effect on osmoporin of K. pneumoniae by molecular docking to further validate the results of in vitro antibacterial property of buffalo colostrum IgG [20]. Osmoporin is a transmembrane pore in the outer membrane of Gram-negative bacteria. It exists as a trimer consisting of chains A, B and C. Chain A of OSM (length 191.8Å) was used for computational analysis (Figure 1).
Figure 1: X-ray crystallographic structure of Outer Membrane Porin (OMP) of K. pneumoniae (PDB ID: 1OSM).
The docked complexes of FDVWGTGT, IVKPGASV, IKSRFI and FIFPP showed strong interaction with the residues Phe 189, Thr 229, Tyr 231, Phe 257. These were imported in XP visualiser and corresponding docking poses were viewed in workspace and compared with the standard drug ampicillin (PDB ID: 4GCP). The overall dock scores (GScore) for the 5 ligands 4GCP, FDVWGTGT, IVKPGASV, IKSRFI and FIFPP were -4.9, -4.8, -4.7, -4.6 and -4.3 respectively (Table 3).
|
Table 3: Docking of 1OSM with IgG peptides from XP method. |
|||||
|
Ligandsa |
G Score |
Lipophilic |
H-bond |
Electrostatic |
Binding energy (kJ mol-1 ) |
|
4GCP_chainA |
-4.9 |
-3.0 |
-1.4 |
0.1 |
-6.12 |
|
FDVWGTGT |
-4.8 |
-4.6 |
-0.8 |
-0.3 |
-5.89 |
|
IVKPGASV |
-4.7 |
-5.2 |
-0.7 |
-0.3 |
-5.56 |
|
IKSRFI |
-4.6 |
-4.5 |
-0.7 |
-0.1 |
-5.24 |
|
FIFPP |
-4.3 |
-4.3 |
-0.9 |
-0.1 |
-4.26 |
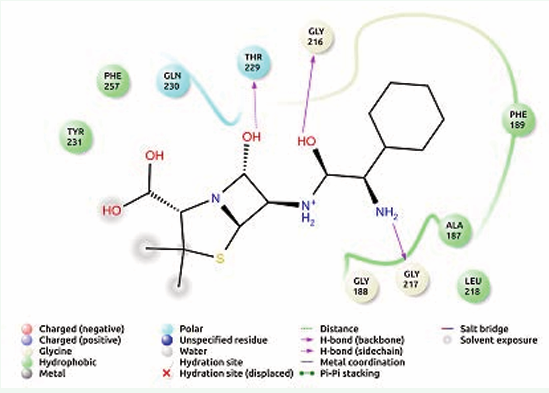
Hydrogen bond interactions in ampicillin were commonly found with Gly 216, Gly 217, Thr 229 and hydrophobic interaction with Ala 187, Phe 189, Leu 218, Tyr 231, Phe 257 (Figure 2).
Figure 2: Receptor ligand interaction of ampicillin with OMP of K. pneumoniae showing interactive amino acid residues in the grid.
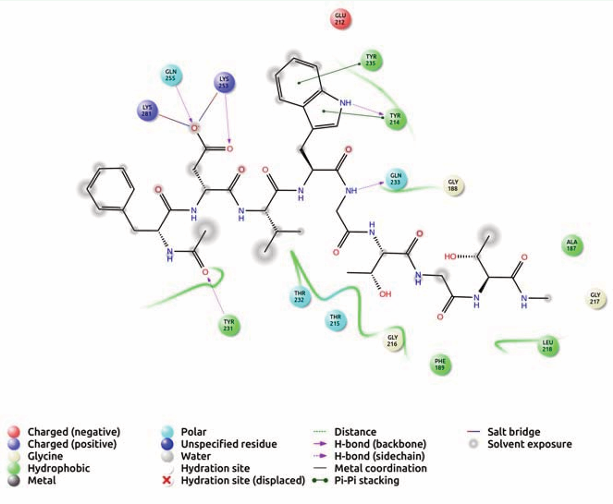
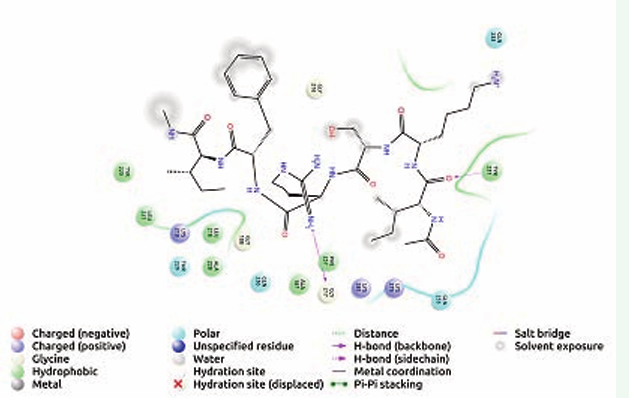
The IgG peptide, FDVWGTGT hydrogen bonded with Tyr 214 and Gln 233 while hydrophobic bonding with Ala 187, Phe 189, Tyr 214, Leu 218, Tyr 231, Tyr 235 (Figure 3) was evident.
Figure 3: Receptor ligand interaction of FDVWGTGT with OMP of K. pneumoniae showing interactive amino acid residues in the grid.
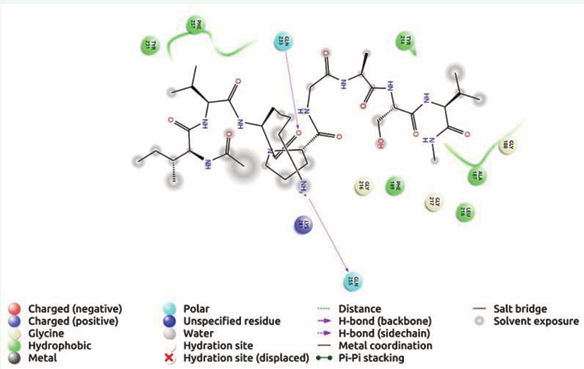
Similarly, IVKPGASV, hydrogen bond interaction showed with Gln 233, Gln 255 and hydrophobic interaction with Ala 187, Phe 189, Leu 218, Tyr 231, Tyr 214, Phe 257 (Figure 4) and IKSRFI,
Figure 4: Receptor ligand interaction of IVKPGASV with OMP of K. pneumoniae showing interactive amino acid residues in the grid.
hydrogen bond interacted with Gly 217, Tyr 231 and hydrophobic with Ala 187, Leu 218, Tyr 220, Leu 227, Ala 228, Tyr 231, Phe 257 (Figure 5).
Figure 5: Receptor ligand interaction of IKSFRI with OMP of K. pneumoniae showing interactive amino acid residues in the grid.
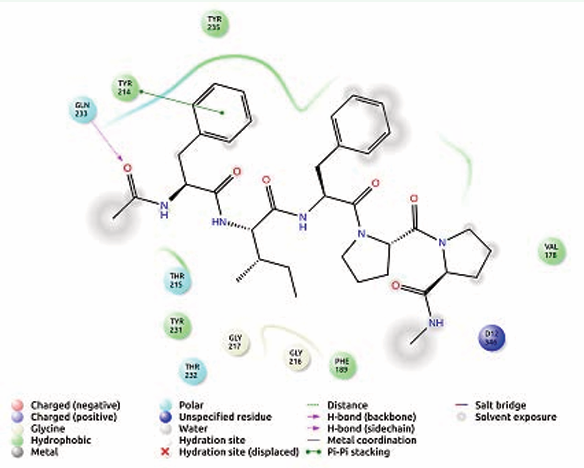
In FIFPP, Gln 233 involved in hydrogen interaction and hydrophobic interaction was found with Val 178, Phe 189, Tyr 214, Tyr 231 and Tyr 235 [Figure 6].
Figure 6: Receptor ligand interaction of FIFPP with OMP of K. pneumoniae showing interactive amino acid residues in the grid.
Some of the interactive amino acid residues were found similar between ampicillin and selected peptides. The Gscore of IgG peptides were found closer to the score of standard antibiotic ampicillin suggesting their probable interaction to the drug target. Further the lipophilic, electrostatic interactions and binding energy among the ligand and the receptor residues were also shown in Table (3).
Molecular modeling has proven increasingly important in complementing in vitro experimental results. Osmoporins are the transmembrane channel which allows diffusion of antibiotics and many other substances and is considered as an ideal target for developing a drug against several bacterial infections [25]. Typically, diffusion of IgG peptides depends on multiple electrostatic interactions, hydrogen bonds and hydrophobic interaction that engage chemical groups on the antibiotic and amino acids of the constriction zone of the porin. The preliminary study carried out using ovine β- casein hydrolyzate after pepsin, trypsin and chymotrypsin digestion was used as a biosensor for quorum sensing molecules and it indicated that the antimicrobial peptides from ovine β-casein may interfere with cell-signaling in bacterial populations [26]. Earlier studies indicate OmpK35 and OmpK36 provide a channel that allows a wide range of antibiotics to penetrate the K. pneumoniae cell wall. These studies have shown that OmpK35 allows for more efficient penetration of cephalosporin than OmpK36 does. Similarly, the IgG peptide interaction with Omp may be new promising therapeutic molecules to treat infections caused by Klebsiella.
CONCLUSIONS
Immunoglobulins are becoming major target oriented biotherapeutics to treat an array of human diseases. IgG are the fastest growing class of therapeutic drugs, because of their high specificities to target cells. Proteomic analysis of buffalo colostrum immunoglobulin has only recently become feasible with the ability to generate peptide motifs of heavy and light chains. Sequences still present significant challenges for mass spectral interpretation due to the frequency of interspersed variable and conserved amino acid sequences within the same peptides. The observation of similar sequence properties in bovine, mouse and human data sets indicates that these are intrinsic features of immunoglobulin primary structure which should be accounted for in any proteomic analysis of antibody repertoire, regardless of species. Moreover, antibodies can act as guides in the quest for small molecules (peptides) that have the ability to modulate protein–protein interactions, which have traditionally only been considered to be tractable targets for biological drugs. Hence, future studies are necessary to determine a safe and effective therapeutics or vaccine/s.
ACKNOWLEDGEMENTS
Authors thank SAIF, IIT, Bombay for nanoLC-MS/MS facility, Schrodinger software, IOE instrumentation facility to carry out this study. Mamatha Bhanu thank University Post-graduate cell for the award of fellowship.