In silico Study of Curcumin Potentiality for IRE1 Oncoproetin Inactivation: A Hypothesis
- 1. Department of Botany, Banaras Hindu University, Varanasi-221005, India
ABSTRACT
The accumulated misfolded or unfolded proteins due to cellular stress and oxidative stress induce endoplasmic reticulum stress, which further activates an unfolded protein response (UPR). ER stress is usually maintained at elevated levels in cancer cell in comparison to normal cells with altered metabolism in cancer. Here, we investigated using molecular docking whether curcumin has interactive properties with IRE-1 in cancer cells as a down regulator. The results from molecular docking between curcumin and IRE1 showed that the curcumin could down regulate the human IRE1 oncoprotein expressing in stress condition.
KEYWORDS
• Curcumin
• IRE1
• Molecular docking
• Stress
CITATION
Srivastava AK, Singh D, Roy BK (2017) In silico Study of Curcumin Potentiality for IRE1 Oncoproetin Inactivation: A Hypothesis. J Bioinform, Genomics, Proteomics 2(1): 1012.
INTRODUCTION
The endoplasmic reticulum is responsible for folding secretory proteins. During homeostasis the folding capacity of the ER and the level of secretory protein synthesis are balanced. Although, if protein synthesis eclipses folding capacity, the level of misfolded protein is sensed and a recovery mechanism is activated, termed the unfolded protein response (UPR). Foremost, UPR signaling reduces protein translation with increasing the ER lumen volume and protein folding capacity, however if homeostasis cannot be restored the UPR signals for the cell to undergo apoptosis [1]. The most evolutionarily conserved UPR pathway is the IRE1- bZIP pathway [2]. The IRE1 resides in the ER membrane, has an N-terminal stress sensing domain in the ER lumen and a cytoplasmic kinase/ribonuclease domain are connected by a single-pass transmembrane helix [3]. The luminal domains dimerise in ER stress taking the cytoplasmic domains together [4]. Further IRE1 undergoes autophosphorylation developing a specific endoribonuclease activity, which hydrolyses 2 stem loops in the pre-mRNA for a bZIP transcription factor [4]. Religation of the RNA causes in a frame shift, and subsequently an active transcription factor is translated, HAC1 in yeast and XBP1 in metazoans [5] that increases transcription of UPR target genes [1]. Recent evidence has explained the UPR activation linking to numerous diseases. The pro-survival IRE1-XBP1 pathway plays a vital role in disease development for inflammatory bowel diseases, metabolic disorders and cancers [6]. The targeting of the IRE1-XBP1 pathway in myeloma and the recent discovery of a key function for this pathway in the progression of aggressive, triple negative breast cancer has revealed that IRE1 has become the focus of several drug discovery programs [7]. A detailed understanding of the IRE1 activation mechanism in humans will accelerate therapeutic development. Therefore present studies focused on a component curcumin as a pleotropic nature is derived from medicinal plant Curcuma longa and are being used against several hazardous diseases. In particular, curcumin, a phenolic compound has been recognized as an antioxidant by scavenging radicals and regulating antioxidant responsive elements and it induces apoptosis via the induction of the cellular stress to the ER in several cancer cells [8]. So, we strived to find interactive potential of curumin with IRE1 to disrupt its natural integrity causing for several diseases including cancer.
EXPERIMENTAL METHODS
Retrieving of 3D structure of compounds
The traditional Indian medicine curcumin (PubChem id: CID_969516) was downloaded in standard data file (SDF) from the database PubChem (www.pubchem.ncbi.nlm.nih. gov) for computational study. The crystal structure of human IRE1 (PDB ID: 5hgi) protein retrieved from protein data bank) has been reported in the literature which possesses different confirmations [9].
Molecular Docking
Docking study of each leading compounds was performed using Patch Dock online server [10]. The number of solutions with their score, area, and six-dimensional transformation space was obtained. These results were used for further refinement and rescoring of 1000 top scoring complexes using Fire Dock [10]. The top ten scoring molecules of each crystal structure were refined automatically by Fire Dock and represented the energy involvement in complex formation as global binding energy, attractive vander Waals, and hydrogen bond energies. Each complex of FireDock was ranked on the basis of minimum global binding energy. Next step, Discovery Studio 4.5 Client [11] for determining the mode of interaction between the receptor and ligands was used.
RESULTS AND DISCUSSION
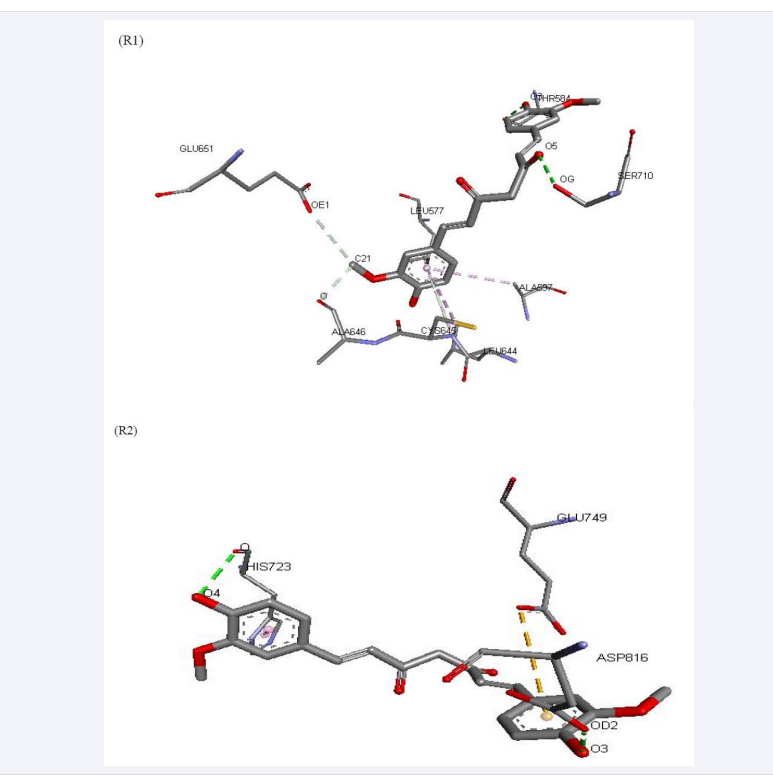
Present studies explain that the curcumin has interactive properties with IRE1 oncoprotein expressed stress condition (Figure 1).
Figure 1: Visualization of interactive molecules of curcumin with strongest (R1) and lowest complexation to IRE1 (R10).
After flexible molecular docking, we found total 10 best solutions by Fire Dock in which rank1 had strongest complexation (Figure 1) and weakest interaction in rank10 (Figure 1) on the basis of global binding energy. Rank1 represents the binding mode of curcumin with residues (THR584, SER710, GLU651, CYS645, LEU644, ALA597, ALA646, and LEU577) with highest binding energy (-35.94 kcal/mol) and Full Fitness (-4318.09 kcal/mol) (Table 1) whereas binding modes of residues (HIS723, GLU749 and ASP816) in rank10 had lowest binding energy (-7.61 kcal/mol) with Full Fitness (-4306.96 kcal/mol) (Table 1). However, spontaneous interactions of curcumin with IRE1 in both of complexes (R1 and R10) exhibited indicating that curcumin had potential to disrupt the natural integrity of IRE1 of any confirmations [12] also studies explained the curcumin promotes ER stress-mediated apoptosis in cancer through increase of cell type-specific ROS. The present study showed that the curcumin has good interactive properties with IRE1 protein and suggested to be potential inhibitor of IRE1 oncoprotein expressed in stress condition. Hence, curcumin could be approached as therapeutic agents against stress proteins causing for cancer development.
Table 1: Inherent free binding energy (kcal/mol) of curcumin-IRE1 complexes.
| Rank | Full Fitness (kcal/mol) | Estimated G (kcal/mol) | Interactive molecules |
| 1 | -4318.09 | -35.94 | UNK1:O3-THR584, UNK1:O5-SER710:OG |
| UNK1:C21-GLU651:OE1, UNK1-CYS645 | |||
| UNK1-LEU644, UNK1-ALA597 | |||
| UNK1:C21-ALA646:O, UNK1-LEU577 | |||
| 10 | -4306.96 | -7.61 | UNK1:O4-HIS723:O, UNK1-HIS723 |
| UNK1-GLU749:OE1, UNK1:O3-ASP816:OD2 |