Methodologies for Ascertaining the Anti-Cancer Potential of DNA G-Quadruplex stabilizers
- 1. Department of Biosciences, Manipal University, India
ABSTRACT
Found mainly at the telomeric, promoter, ribosomal RNA and regulatory regions in genome, G4 quadruplexes are guanine-rich region that cause DNA strands to get folded into planar quadruplex with the help of Hoogsten hydrogen bonds and stabilization by metal ions such as Na+ and K+.G-quadruplexes impact several cellular processes such as telomere maintenance, replication, transcription, recombination and repair. These G-quadruplex structures present natural impedance to enzymes and protein factors related to mentioned cellular processes. Under normal physiological conditions in healthy cells, a number of proteins (called helicases such as PIF1, BLM, WRN, FANCJ) have ability to unwind or resolve these G-quadruplex structures, and thus, diminish possible damage to DNA and ultimately maintain secure genome stability and integrity. In this way, normal healthy cells escape pathological consequences such as cancer. However, stabilization of G-quadruplex structures by certain chemically synthesized molecules also provides a way to induce synthetic lethality in cells, which can also be selectively beneficial in inhibiting replication, transcription, recombination and repair in cancer cells which ultimately will lead to cancer cells’ death.
KEYWORDS
• G4 structures
• G-quadruplex
• Cancer
• G4 binders
• Stabilizers
CITATION
Dakal TC (2018) Methodologies for Ascertaining the Anti-Cancer Potential of DNA G-Quadruplex stabilizers. J Bioinform, Genomics, Proteomics 3(1): 1028
ABBREVIATIONS
BLM: Bloom Syndrome RecQ like Helicase; DOG1: Deletions of G-rich DNA; FANCJ: Falconi Anemia Complementation Group J; PIF1: PIF 5’-3’ DNA Helicase; WRN: Werner Syndrome RecQ like Helicase
INTRODUCTION
The propensity of G-rich regions in genome to self-associate and form higher-order tetramolecular (four-stranded) DNA and RNA structures is known since 1960s [1]. G-rich region in genome such as G3+--N1-7-G3+-N1-7--G3+--N1-7—G3+ (G4 motifs)are supposed to form G4 structures, formally known as G-quadruplexes [2]. G-quadruplexes represent tetrads of hydrogen-bonded guanine bases stabilized by Hoogsten hydrogen bonds and metal ions such as Na+ and K+ where the building blocks are G-quartets that stack on top of one another (analogous to Watson–Crick base pairs) and form topologically polymorphic intra- and inter-molecular tetrad helical structures. So far, more than 375,000 G quadruplexes have been mapped in human genomeand in thousands in Saccharomyces cerevisiae genome also [3-6]. G quadruplexes are not present all over the genome but are predominantly enriched at telomeres, rDNA, nucleosome depleted, regulatory, promoter and 5′ untranslated regions (5’ UTRs) of highly transcribed genes, in particular those linked with tumorigenesis and somatic copy number amplifications, such as MYC [3-7]. There existsa number of striking evidences in relation to involvement of G4 structures in regulation of telomere maintenance, replication, transcription, translation, recombination and repair that suggest that G4 contribute to and regulate genetic and epigenetic stability of the genome. With the advent of specialized antibodies directed against G-quadruplexes (such as BG4 and 1H6)it is possible to ascertain the number of DNA damage foci in different cell cycle phases and it has been found that DNA damage foci significantly increased during S-phase [8].
Being highly thermally stable, formation of G-quadruplexes impose natural impedance to several cellular DNA transactions processes such as replication, transcription, translation, recombination and repair [2,9]. It is essential to resolve the G quadruplexes in order to allow normal DNA replication, transcription, translation, recombination and repair.There are mounting evidences that some specialized DNA polymerases and helicases resolve G-quadruplexes at replication fork site and allow normal replication of G quadruplexes and thereby prevent genetic and epigenetic instability [10,11]. The possible role of G-quadruplex helicases in DNA replication came from the studies on genome instability syndromes caused by functional mutations in one or more such DNA helicases. For instance, in Caenorhabditis elegans, as a consequence of functional mutation in the DOG-1 gene, deletions accumulate in regions harboring pG4s in the genome and dog1 deficient C. elegans experience increased stalling at G-quadruplexes during replication [12,13]. Studies on other helicases, as for example FANCJ, WRN, BLM and Pif1, support and extend these findings that is functional defects in any of the associated cellular pathway such as replication, transcription, translation, recombination and repair [13]. FANCJ as well as Pif1 or BLM helicase primarily act at internal genomic regions and are required for genome integrity [10,11]. As for example, in humans depletion of the FANCJ/ BRIP1 helicase causes persistent stalling at G-quadruplexes during replication suggesting indispensable role of this helicase in resolving tetramolecular G-quadruplexes at replication fork site and FANCJ performs this independently of the classical Fanconianemia pathway [13]. On contrast, WRN, a helicase associated with Werner syndrome, acts mainly at telomeric regions. RPA is also associated with unfolding of G-quadruplex structures formed by telomeric DNA sequences, a function important for telomere maintenance [14]. Functional mutations in WRN helicase causealtered replication of the telomeric DNA, consistent with Werner syndrome patients having mutations in the WRN helicasethat results in premature senescence and accelerated telomere shortening. In Saccharomyces cerevisiae, Pif1 deficiency led to replication being slowed down in the vicinity of pG4s andoccurrence of DNA breakage at these sites suggesting the role of Pif1 in resolving G-quadruplexes in yeasts G-quadruplex associated DNA breakage could be suppressed by the ectopic expression of Pif1 [15,16]. BLM helicase, a RecQ family helicase implicated in Bloom syndrome (an autosomal recessive disorder) and associated with growth retardation and predisposition to cancer. Using single molecule FRET analysis it has been shown that BLM helicase with high specificity interacts with non-canonical intramolecular G-quadruplex structures [17].
Recent emphasis has been directed towards recognition and resolution of G-quadruplex structures in human genome using biochemical, biophysical and structural biology methods to provide valuable insights into transcriptome, proteome and metabolome aspects of normal cells as well as cells in pathological context. Using the essential information obtained from these studies and from high-resolution methods such as X-ray crystallography and NMR, researchers are able to discover and design novel chemical molecules and drugs targeting G-quadruplexes. Several researchers, using different approaches, are able to discover, design and synthesize chemical molecules that can specifically bind to and stabilize G-quadruplex structures that form stably and transiently in genome [18-20]. Stabilization of G-quadruplex structures by these chemical molecules inhibit vital cellular processes such as replication, transcription, translation, recombination and repair leading to cell apoptosis and death. This provides a way to induce synthetic lethality in the cell and could be used as a promising therapy to specifically target cancer cells. This strategy can be coupled with other strategy such as depletion of essential DNA repair proteins/factors using gene deletion or chemical inhibition for further inhibiting DNA repair in cancerous cells which will ultimately lead to increased cancerous cells’ killing. Using the methodology described in this paper, the anti-cancer potential of the chemical G-quadruplex stabilizers can be ascertained at genome, transcriptome, proteome and metabolome level (Figure 1). Once proved successful under in-vitro conditions, the future work can be directed towards designing an appropriate drug delivery system to target G4 stabilizers to specificorgan/tissue under in-vivo conditions. The strategy is still in infancy; however, holds immense potential for promising management of different cancers in future.
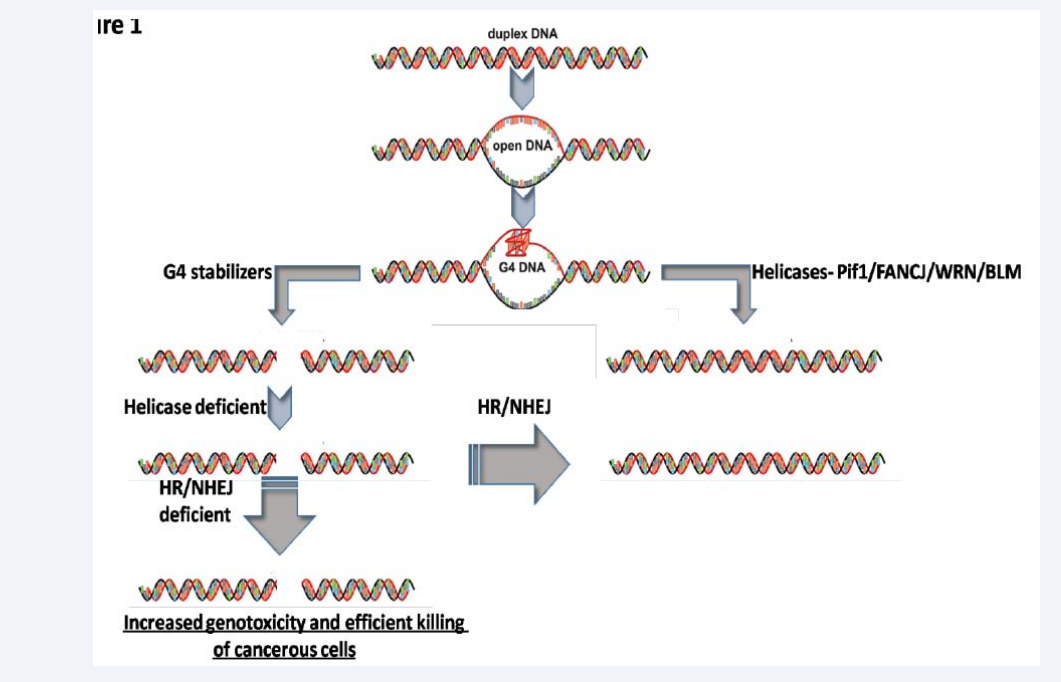
Figure 1: The strategy to induce synthetic lethality in cancer using G4 stabilizers in presence of depletion of key DNA repair proteins/pathways.
DIFFERENT METHODOLOGIES FOR ASCERTAINING THE ANTI-CANCER POTENTIAL OF G-QUADRUPLEX STABILIZERS
Methodologies or experimental strategies one should use to ascertain the anti-cancer potential of chemical stabilizers of G-quadruplex structures depend mainly upon the hypotheses or assumptions one make. In context to cancer following hypotheses become apparently important to be tested.
Hypothesis no. 1: Stabilizers of G4 structures are expected to inhibit the replicative growth of cells and this can be selectively used inhibit tumor cell growth in-vitro. This can be validated using proliferation and apoptosis assay as described below.
Cell proliferation and clonogenic assay: To determine the average rate of population doublings, different cancer cell lines (treated with G4 stabilizers and non-treated)will be plated into petri-dishes with 10 cm diameter in triplicatewith initial cell density of 1.0 × 10(6) cells/dish. After the indicated intervals (0 to 48 h), cells can betrypsinized and counted using the Countess® Automated Cell Counter (Life Technologies) or using Hemocytometer after cell staining with trypan blue. In order to check reproductive death after treatment with G-quadruplex stabilizers, one can perform the clonogenic assay in which the cell lines with an initial cell density of 100-200 cellscan be plated in 6cm diameter plates 24h prior to treatment with and without the G-quadruplex stabilizers and can be incubated for a specified time after which colonies can fixed with glutaraldehyde (6.0% v/v) stained with crystal violet (0.5% w/v) and counted to determine colony forming units.
Stabilization of G4 structures by certain chemically synthesized molecules provides a way to induce synthetic lethality in cells, which can also be selectively beneficial in inhibiting proliferation of cancer cells.
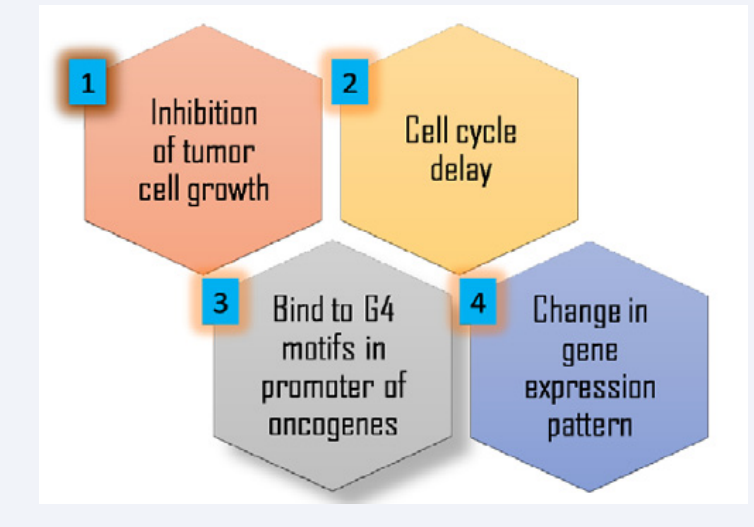
The conditional hypothesis is that in order to act as anti-cancer agents, G4 binders are expected to cause following: (Figure 2).
Figure 2: Methodologies for ascertaining the anti-cancer potential of G-quadruplex stabilizers.
Apoptosis assay
Apoptotic cell death can be measured using one of these: DNA fragmentation (gel electrophoresis), cyt C release (using WB), Bcl-2 expression (WB), change in mitochondrial membrane potential (cell staining), glutathione assay (fluorescence measurement) according to standard protocols suggested by manufacturers.
Immunofluorescence microscopy:IF microscopy using 1H6 and BG4 antibody. 1H6 detects only those G4 structures that are more stably present in the genome [8]. As an alternate, BG4, which has high affinity for both stably and transiently formed intra- and intermolecular DNA G4 structures, can be used.
Hypothesis no. 2
Stabilizers of G4 structures are expected to cause a delay in cell cycle progression through S-phase. This will be validated using cell cycle analysis using BD FACS Calibur as described below.
Cell cycle analysis using flow cytometry: Different cancer cell lines can be synchronized and blocked at the G1 or G1/S border or M-phase. The synchronized cancer cells can be treated with and without G4 stabilizersand can be allowed to grow under normal condition after which cell can be harvested and fixed in ice-cold 70% (v/v) ethanol for 24 h at 4 °C for cell cycle analysis. The cell pellet can be collected by centrifugation at 500×g, resuspended in PBS, stained with a mixture of RNase (10 µg/mL) and PI (50 µg/ mL) in sodium citrate buffer containing 0.5% Triton X-100 for 20 min in the dark. Cell cycle analysis can performed using a BD FACS Calibur™ as per standard procedure.
Hypothesis no 3
Since, stabilization of G4 structures by G4 stabilizers is expected to pull down more G4 structures from a sample of sonicated genomic DNA using Hf2 antibody-Protein A based pull down assay. This will allow mapping of increased number of G4 structures, including that form transiently under only certain physiological conditions.
Hf2 pull down assay: Hf2 antibody expression and purification canbe carried out as previously described with following modifications [21]. Cells can be harvested and thepellets can be resuspended in lysis buffer (25mM Tris-HCl, 100 mMNaCl, 10% glycerol, 1% NP-40 and 10 mMimmidazole) followed by sonication using HisPur Ni-NTA resin (ThermoFisher Scientific,Wilmington, Delaware, USA) following the manufacturer-suggested protocol. Eluted protein can be concentrated using Amicon Ultra-4 Centrifugal Filter and can stored at –20o C in 50% glycerol. Following steps can be as per the protocol suggested by Lam et al. (2013) [21].
Hypothesis no. 4
Since, G4 structures also effect genomic transcription, and therefore, G4 stabilizers are expected to cause a drastic change in gene expression pattern in normal and cancer cells in response to treatment with G4 stabilizers. This can be ascertained using microarray analysis followed by qPCR as described below. This will further enhance our understanding of genes and molecular pathways affected.
Differential gene expression analysis using microarray: G4 structures present in the promoter regions of any gene will influence the transcription of the gene. One canuse Illumina Bead Chip Microarray. The Illumina raw data files can be imported into the Flex Array (ver 1.6.3), a software tool for gene expression data analysis, in which the execution of robust multi-array average (Lumi), which performs the background correction and Robust spline normalization, can be carried out. For subsequent statistical analysis SAM (Significance analysis of microarray), an algorithm implemented in Flex Array, can be used. Genes up-regulated (≥2 fold change) and down-regulated (≤ 0.5 fold change) with high statistical significance (p value <0.01 or less) can be exported as .txt file for further analysis such as volcano plots, venn diagram and heat maps. The gene ontology (GO analysis) and pathway analysis can be done using DAVID.
Validation and quantification of differential gene expression analysis using qPCR: The total cellular RNA will be isolated using RNA easy mini kit (Qiagen; Cat no. 74104) according to the manufacturer’s protocol. RNA concentration can be measured with Nano Drop 2000 spectrophotometer (Thermo fisher Scientific, Wilmington, Delaware, USA). The first-strand cDNA synthesis will be done using 100 ng of total RNA primed with oligo(dT) using native avian myeloblastosis virus reverse transcriptase in the presence of RNase inhibitor RNaseOUT (Thermo fisher Scientific, Wilmington, Delaware, USA; Cat no. 10777-019). The cDNA will be diluted to 10 ng/μ l using RNase-free water. Quantitative real-time PCR (qRT-PCR) amplification reactions will be performed with the Applied Biosystems 7500 RT PCR system (Applied Biosystems), using the PerfeCTa SYBR Green SuperMix Low Rox PCR Kit (Quanta Biosciences; Cat no. 95056). The PCR mix and the conditions will be set appropriately. The threshold numbers (Ct values) will be set within the exponential phase of the reaction and will be used to calculate the relative expression for each gene normalized to either ACTβ (β -actin) or HPRT RNA in each sample (Figure 3).
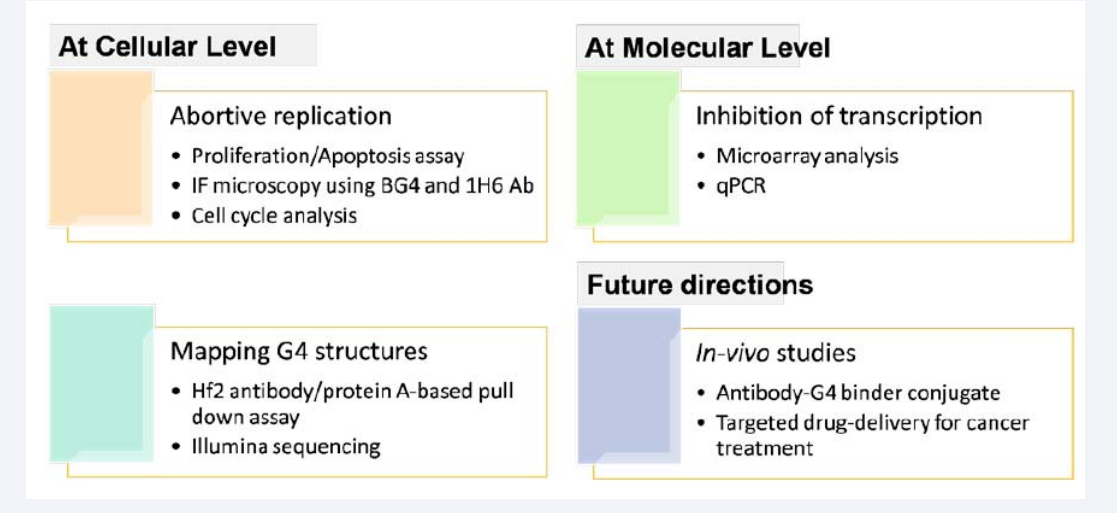
Figure 3: Most important hypotheses and assumptions made in relation to anti-cancer potential of G-quadruplex stabilizers.
However, the corresponding methodologies of each hypothesis can be fully proved once detailed experimental outcomes are obtained.Besides this, the functional consequences of SNPs/mutations in the G4 motifs/G-quadruplexes and in different helicase proteins can be studies based on bioinformatics and structural modelling approach as per the Dakal et al. (2017a, b) [22,23]. Besides this, G-quadruplexes are also preferentially present in rDNA regions and it would be interesting to know if such structures are present in the rDNA regions of some yeasts, in particular of genus Zygosaccharomyces and some other that show hyper-variable rDNA regions along with corresponding genotypic and phenotypic diversity [24-26]. Whether G-quadruplexes in the rDNA region of Zygosaccharomyces and the sequence polymorphism withinthis region explain any roles related genotypic/phenotypic diversity and evolution could be ascertained thereafter [27-31].
CONCLUDING REMARK
The outcome of the proposed study is expected to bring novel insights in relation to G-quadruplexes and its implication in cancer such as a) the cellular, molecular and genetic basis of novel G-quadruplex stabilizers’ based anti-cancer therapy described herein, b) types of cancers that can be treated using the novel G-quadruplex stabilizers’ based anti-cancer therapy described herein, c) discovery of some novel transient (less stable) G-quadruplex structures and their role in regulating gene expression in relation to cancer. The experimental work on verifying reasonability of these experiments will be the future prospects of this paper.Ultimately such study will be useful in assistingthe scientific communityin understanding major impediments related to successful diagnosis and treatment of cancer and also in revising existing therapies for successful management of different cancer types.
ACKNOWLEDGMENTS
I would like to thank Manipal University Jaipur for the facility.