Synthesis of Phenylpiperazine Derivatives of 1,4-Benzodioxan as Selective Tyrosyl-Trna Synthetase Inhibitors
- 1. School of Life Sciences, Shandong University of Technology, Zibo 255049, People’s Republic of China
- 2. Shandong Experimental High School, Jinan 250001, People’s Republic of China
ABSTRACT
Five phenylpiperazine derivatives of 1,4-benzodioxan have been designed, and evaluated for antimicrobial activities. Docking simulations have been performed to position compounds into the Tyrosyl-tRNA synthetase active site to determine their probable binding models. All of the compounds exhibited better antibacterial activities against Gram-positive strains. Interestingly, compound 3c exhibited better antibacterial activities with IC50 values of 0.03 µg/mL against Staphylococcus aureus.
KEYWORDS
• Phenylpiperazine
• 1,4-benzodioxan
• Tyrosyl-TRNA synthetase
• Antibacterial activities
• Molecular docking
CITATION
Xu RF, Liu HY, Sun J, Chen ZW (2018) Synthesis Of Phenylpiperazine Derivatives Of 1,4-Benzodioxan As Selective Tyrosyl-Trna Synthetase Inhibitors. J Bioinform, Genomics, Proteomics 3(1): 1027.
INTRODUCTION
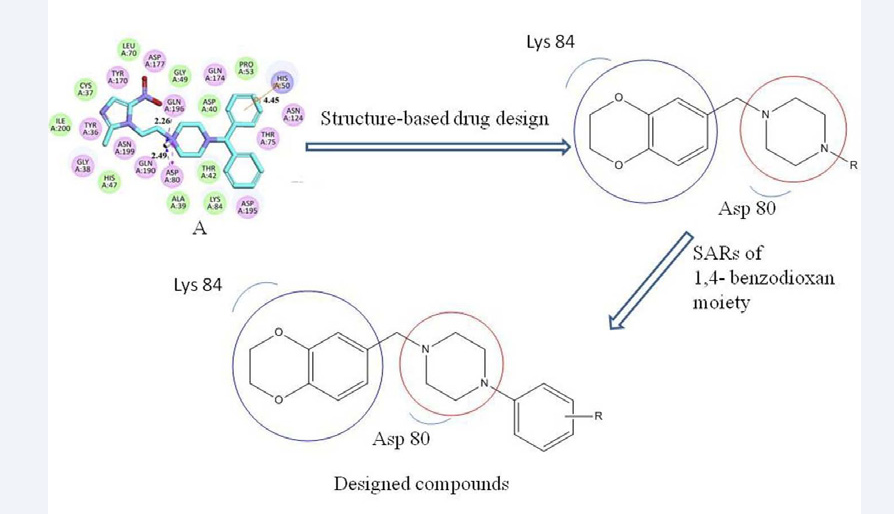
Aminoacyl-tRNA synthetases (aaRSs) are a kind of key enzymes which catalyze the transfer of amino acids to their cognate tRNAs during protein synthesis [1-3]. AaRSs have been proved to be antimicrobial targets [4], and they have been interesting targets in antibacterial drug design [5-7]. As a member of aaRS family, Tyrosyl-tRNA synthetase (TyrRS) are found in all living organisms and plays an important role in protein synthesis [8-12]. TyrRS belong to the class I tRNA synthetase family, which has two highly conserved sequence motifs at the active site, HIGH and KMSKS [13,14]. There are several distinctive differences between bacterial and human TyrRS. These properties suggest that small-molecule inhibitors of TyrRS could be promising drug candidates leading to high selectivity and broad-spectrum antibacterial agents [15,16]. Phenylpiperazine derivatives have showed broad varieties of biological activities especially antimicrobial activity. As a continuous work of our previous work [17], we have researched a series of phenylpiperazine derivatives as potential TyrRS inhibitors. Besides, it is reported that 1,4-benzodioxan afforded a new scaffold for small molecular antibacterial activity [18,19]. Thus our main objective is to design novel phenylpiperazine derivatives of 1,4-benzodioxan as specific inhibitors of TyrRS in the hope that these molecules may be further explored as powerful and novel antibacterial lead-candidates. Our strategy is intended to obtain potent antibacterial activity with selective inhibition of TyrRS using traditional medicinal chemistry techniques motivated by the comparative modeling of TyrRS complexed with A in the available pharmacophore (Figure 1).
Figure 1: Design strategy of the target compounds.
Therefore, we combined two scaffolds and made the molecular docking (Figure 2).
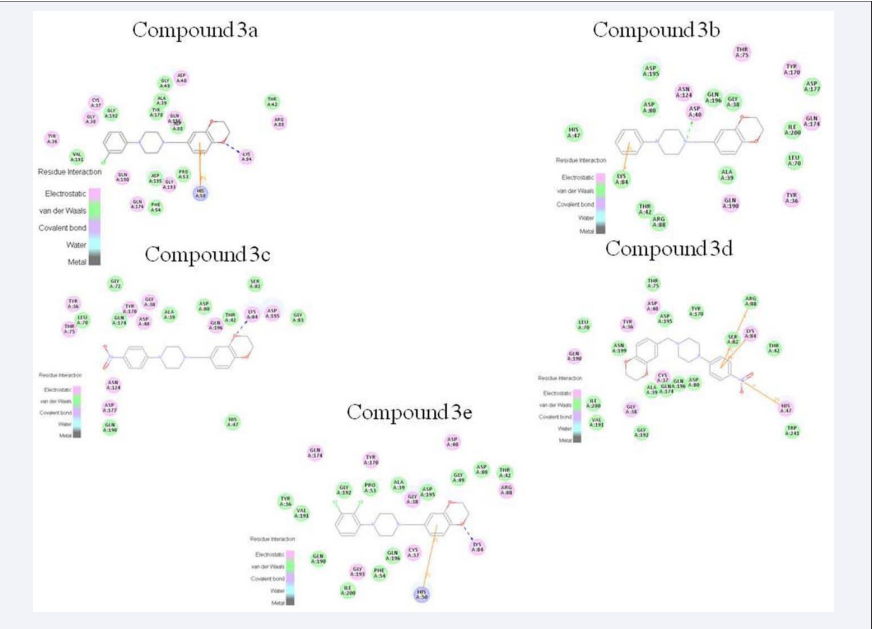
Figure 2: 2D molecular docking modeling with 1JIJ.
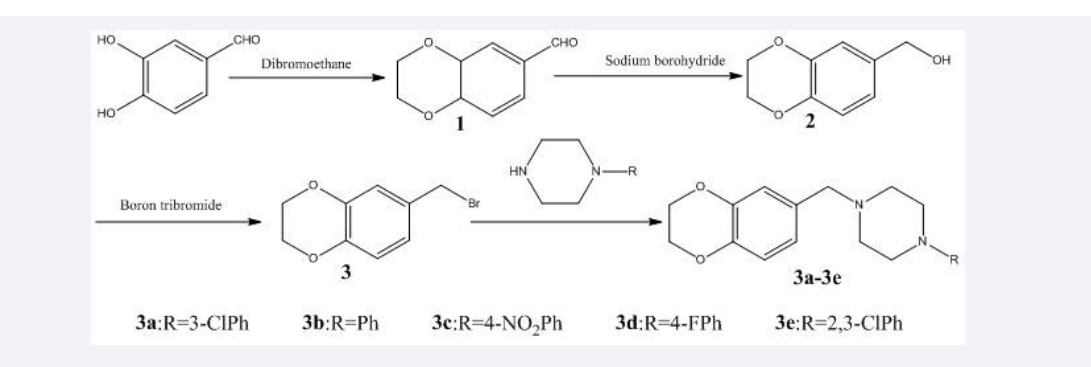
In the binding model, compounds 3a-3e were bound to the TyrRS with hydrogen interaction bonds, π-cation bonds and π-π bonds. Therefore, these preliminary analysis served as a stimulant to synthesize these phenylpiperazine derivatives containing 1,4-benzodioxan skeleton 3a-3e. (Scheme 1).
Scheme 1: General synthesis of compounds 3a–3e.
RESULTS
Antibacterial activities
The IC50 of compounds 3a-3e against these bacterial strains are tested by MTT method. Based on the data obtained (Table 1), we found that Compounds 3a-3e possess high selectivity to exhibit better antibacterial activity against Gram-positive bacteria; compound 3c exhibited better antibacterial activities with IC50 values of 0.03 µg/mL against Staphylococcus aureus. Compound 3d displays antibacterial activity with IC50 values of 0.07µg /mL against Bacillus subtilis. Based on the data, we can concluded that the substituent at different positions led to different activity, and para-substituted compounds have the best activities.
Crystal structures of compounds 3a-3e
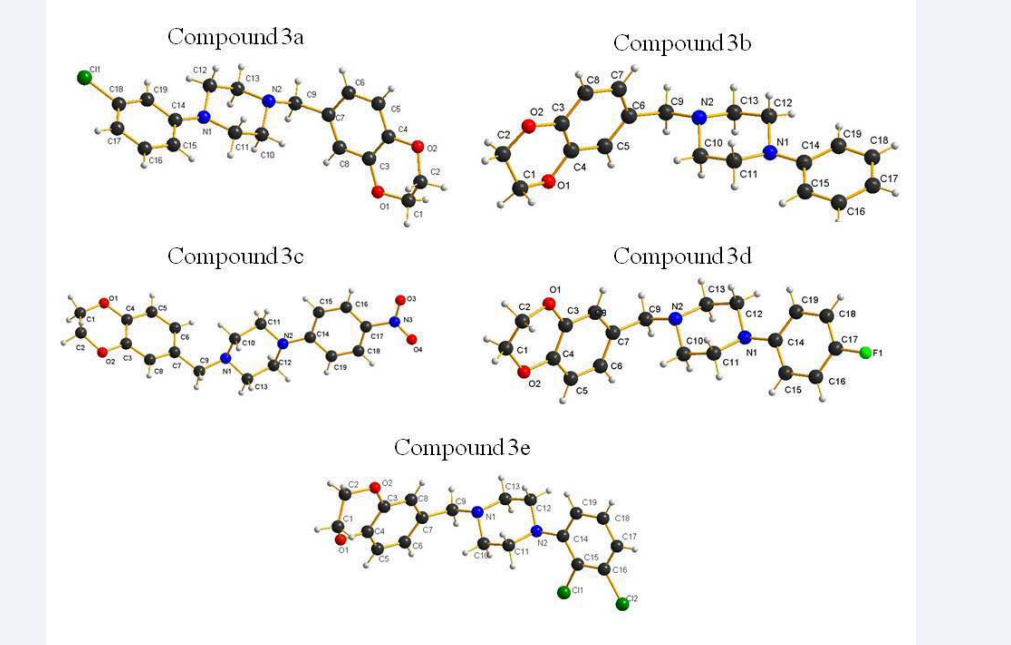
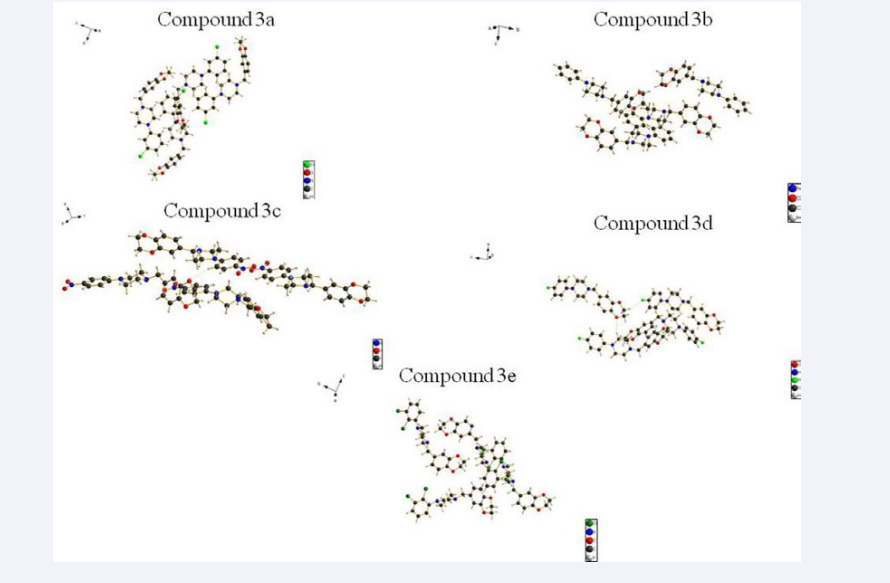
Crystals of five compounds were obtained from methanol solution. Figure (3) shows a perspective view of the monomeric unit with the atomic numbering scheme, Figure (4) depicts the intra-molecular and inter-molecular hydrogen bonds.
Figure 3: Molecular structures of the title compounds with atomic numbering scheme.
Figure 4: Crystal packing of the title compounds.
Crystallographic data, details of data collection and structure refinement parameters are listed in Table (2). Single crystals of 3a (0.21 mm×0.21 mm×0.18 mm), 3b (0.21 mm×0.21 mm×0.18 mm), 3c (0.21 mm×0.21 mm×0.19 mm), 3d (0.21 mm×0.21 mm×0.18 mm), 3e (0.21 mm×0.21 mm×0.19 mm), were mounted on a D-8 venture diffractometer equipped with graphite-monochromated MoKα (k = 0.71073 Å) radiation.
For 3a, a total of 16,546 reflections were collected, of which 3229 were unique with Rint = 0.054 and 1947 observed reflections with I > 2σ(I) were used in the succeeding structure calculations. The final cycle of refinement of full matrix least-squares was converged to R = 0.0593 and wR = 0.1649. The highest and lowest residual peaks in the final difference Fourier map are 0.40 and -0.32 e/Å3 , respectively. For 3b, a total of 15,962 reflections were collected, of which 3076 were unique with Rint = 0.060 and 1669 observed reflections with I > 2σ(I) were used in the succeeding structure calculations. The final cycle of refinement of full matrix least-squares was converged to R = 0.0537 and wR = 0.1416. The highest and lowest residual peaks in the final difference Fourier map are 0.24 and -0.17 e/Å3 , respectively. For 3c, a total of 30,921 reflections were collected, of which 6622 were unique with Rint = 0.045 and 4026 observed reflections with I > 2σ(I) were used in the succeeding structure calculations. The final cycle of refinement of full matrix least-squares was converged to R = 0.0557 and wR = 0.1497. The highest and lowest residual peaks in the final difference Fourier map are 0.29 and -0.25 e/Å3 , respectively. For 3d, a total of 16,425 reflections were collected, of which 3163 were unique with Rint = 0.029 and 2693 observed reflections with I > 2σ(I) were used in the succeeding structure calculations. The final cycle of refinement of full matrix least-squares was converged to R = 0.0417 and wR = 0.1089. The highest and lowest residual peaks in the final difference Fourier map are 0.22 and -0.27 e/Å3 , respectively. For 3e, a total of 17,610 reflections were collected, of which 3469 were unique with Rint = 0.027 and 2781 observed reflections with I > 2σ(I) were used in the succeeding structure calculations. The final cycle of refinement of full matrix least-squares was converged to R = 0.0411 and wR = 0.1146. The highest and lowest residual peaks in the final difference Fourier map are 0.35 and -0.20 e/Å3 , respectively.
Molecular docking study
Automated docking studies were carried out using Discovery Studio (version 3.5) as implemented through the graphical user interface DS-CDocker protocol.
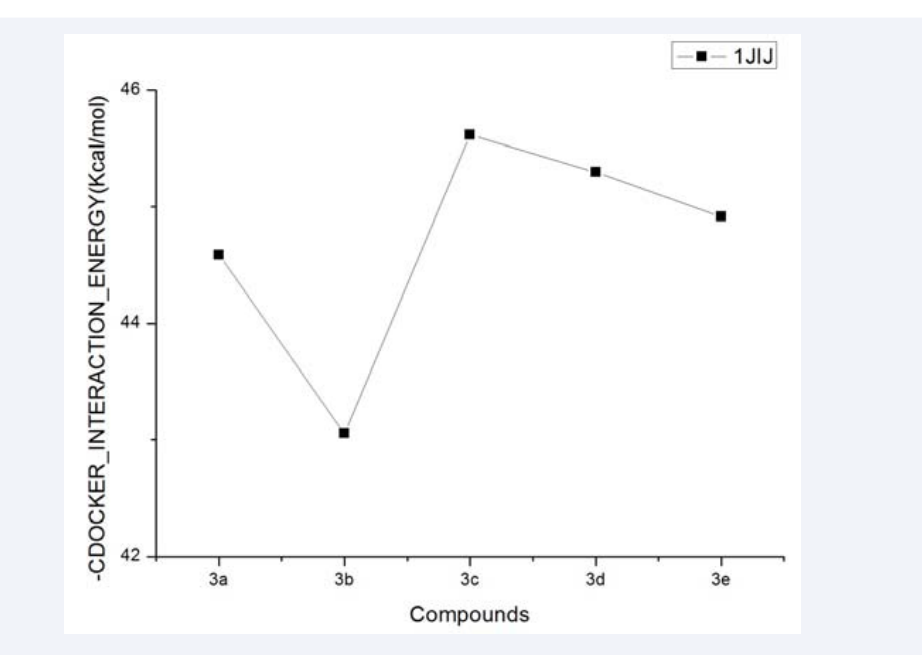
The results have been plotted as a line-scatter graph and presented in Figure 5.
Figure 5: The CDOCKER_INTERACTION_ENERGY (kcal/mol) obtained from the docking study of all synthesized compounds by the CDOCKER protocol (DS 3.5, Accelrys, Co. Ltd).
We could see that compound 3d and 3e have the lower interaction energy than the others, which is consistent with the antibacterial activities.
Table 1: Inhibitory activity (IC50) of the synthetic compounds against bacteria.
| Compound | IC50 (µg/mL) | |||
| Staphylococcus aureus | Bacillus subtilis | Escherichia coli | Pseudomonas aeruginosa | |
| 3a | 1.78 | 14.22 | >50 | >50 |
| 3b | 2.15 | 11.18 | >50 | >50 |
| 3c | 0.03 | 0.28 | >50 | >50 |
| 3d | 0.17 | 0.07 | >50 | >50 |
| 3e | 3.58 | 5.12 | >50 | >50 |
Table 2: Crystallographic data, details of data collection and structure refinement parameters.
| Compound | 3a | 3b | 3c | 3d | 3e |
| Empirical formula | C19H21ClN2 O2 | C19H22N2 O2 | C19H21N3 O4 | C19H21FN2 O2 | C19H20Cl2 N2O2 |
| Formula weight | 344.83 | 310.39 | 355.39 | 328.38 | 379.27 |
| Crystal system | Monoclinic | Monoclinic | Monoclinic | Orthorhombic | Monoclinic |
| Space group | P21/c | P21/c | P21/n | P212121 | P21/n |
| a (Å) | 8.9539(16) | 8.9980(7) | 10.0927(5) | 9.7090(11) | 9.6197(6) |
| b (Å) | 17.099(3) | 16.4968(12) | 9.9642(6) | 11.1305(13) | 14.5024(9) |
| c (Å) | 11.0759(19) | 11.0039(9) | 34.6853(18) | 15.4361(18) | 13.7924(8) |
| α(º) | 90 | 90 | 90 | 90 | 90 |
| β(º) | 94.156(6) | 94.406(3) | 94.073(2) | 90 | 109.369(2) |
| γ(º) | 90 | 90 | 90 | 90 | 90 |
| V(Å) | 1691.3(5) | 1628.6(2) | 3479.3(3) | 1668.1(3) | 1815.26(19) |
| Z | 4 | 4 | 8 | 4 | 4 |
| D calc/g cm¯³ | 1.354 | 1.266 | 1.357 | 1.308 | 1.388 |
| θRange(º) | 2.2, 25.8 | 2.2, 25.7 | 2.1, 25.7 | 2.3, 25.7 | 2.1, 25.8 |
| F(000) | 728 | 664 | 1504 | 696 | 792 |
| Reflections collected/unique | 16,546/3229 | 15,962/3076 | 30,921/ 6622 | 16,425/3163 | 17,610/ 3469 |
| Data/restraints/parameters | 1947/0/ 217 | 1669/0/ 208 | 4026/0/ 469 | 2693/0/ 217 | 2781/0/ 226 |
| Absorption coefficient (mm¯¹ ) | 0.240 | 0.083 | 0.097 | 0.093 | 0.373 |
| R1 /WR2 [I > 2σ(I)] | 0.054 /0.1649 | 0.060/ 0.1416 | 0.045/ 0.1497 | 0.029 /0.1089 | 0.027/ 0.1146 |
| R1 /WR2 (all date) | 0.0593/0.1649 | 0.0537/ 0.1416 | 0.0557/0.1497 | 0.0417/0.1089 | 0.0411/ 0.1146 |
| GOOF | 1.041 | 1.014 | 1.022 | 1.052 | 1.049 |
CONCLUSION
In conclusion, a series of phenylpiperazine derivatives of 1,4-benzodioxan 3a-3e were synthesized, structure characterization, molecular docked and tested for their inhibitory activities against E. coli, P. aeruginosa, B. subtilis and S. aureus. According to the data presented in Figure 2, the compounds were nicely bound to the TyrRS with hydrogen interaction bonds, π-π bonds and π-cation interactions. Especially, compound 3a and 3e were primely bound to LYS 84 with H-bond interaction, and bound to the HIS 50 with π-π interaction. Compound 3b were bound to ASP 40 with H-bond interaction, and bound to the LYS 84 with π-cation interactions. Furthermore, the docking studies of compounds reveals that 1,4-benzodioxan as one scaffolds was primely bound to the LYS 84. Thus, the introduction of hydrogen bonds makes the corresponding compounds possess high selectivity antibacterial activities. From the theoretical perspective, we could conclude that the target compounds were appropriately positioned into the TyrRS active site.
However, according to the data presented in Table (1), compounds 3a-3e possess high selectivity to exhibit better antibacterial activities against two Gram-positive strains. Besides, compound 3c displays antibacterial activity against S. aureus from the close inspection of the data, the activities of the target compounds are consistent with the theoretical results.
SUPPLEMENT FILES
Experimental protocol is in the supplement files.
REFERENCES
2. Arnez JG, Moras D. Trends in biochemical sciences. 1997; 22: 211-216.
6. Ibba M, Söll, D., Annual review of biochemistry, 2000; 69: 617-650.
14. Sun J, Lv PC, Zhu HL. Expert Opin Ther Pat. 2017; 27: 557-564.