In silico Analysis of Protein
- 1. Applied Science Division, Indian Institute of Information Technology, India
Abstract
Proteins are inimitable as principal functional agent of living system. Therefore, comprehension of protein sequence and structure and its correlation with its function is equivalent to deciphering almost all of fundamental features of any biological/living system. A treasure of In silico tools is accessible for analysis of protein. Understanding and regeneration of protein function requires comprehension of reliance between protein sequence and its structure, its localization in cell and its interaction with other functional partners. This review provides an insight for various tools for In silico analysis of protein.
Citation
Singh N, Upadhyay S, Jaiswar A, Mishra N (2016) In silico Analysis of Protein. J Bioinform, Genomics, Proteomics 1(2): 1007.
Keywords
• Protein analysis
• In Silico
• Function
• Sequence
• Tools
ABBREVIATIONS
UNIPROT: Universal Protein Resource; FASTA: FAST Alignment; BLAST: Basic Local Alignment Search Tool; Blastn: Nucleotide Basic Local Alignment Search Tool; Blastp: Protein Basic Local Alignment Search Tool; psi-BLAST: Position-Specific Iterated Basic Local Alignment Search Tool; SOPMA: Self Optimized Prediction Method with Alignment; GOR4:Garnier-Osguthorpe-Robson 4; I-TASSER: Iterative Threading Assembly Refinement; RAMPAGE: Rama chandran Plot Assessment; CASTp: Computer Atlas of Surface Topology of Proteins; T-COFFEE: Tree-based Consistency Objective Function For alignment Evaluation
INTRODUCTION
Tools for In silico analysis of protein are of major significance for making use of data for development of drug and health care. Proteins are complex macromolecules which have essence role to cellular activity. They increase biological processes or reactions by expression of catalytic activity. Thus, analysis and knowledge of protein is substantial for determining the protein function and to understand their role and mechanism in human body [1].
Protein sequence retrieval
Sequence of protein is retrieved from UniProt (Universal Protein Resource) which is a freely accessible database which contains data of proteins. The required protein sequence is retrieved through their accession number and it is in FASTA format. This sequence is further utilized for primary and secondary structure analysis. To obtain sequence similarity of query protein sequence with the known protein structure BLAST (Basic Local Alignment Search Tool). This tool compares protein or nucleotide query sequence to database of sequences and evaluates analytical importance of matches tool is used. Blast is group of programs including blastn (searches DNA sequences against DNA database) ,blastp (for a given protein query sequence returns similar sequence from database of protein sequences), psi-blast (it establishes distant relationship between proteins), blastx (comparison of six-frame translation product of query sequence of nucleotide against database of protein sequence) , tblastx (comparison of six-frame translation product of query sequence of nucleotide against database of nucleotide sequence) andtblastn (comparison of protein query sequence against six reading frames of database of nucleotide sequence) [2].
Primary structure prediction
The primary structure analysis of protein and physicochemical depiction is done using ProtParam tool from ExPasy (Expert Protein analysis system) (Figure 1).
Figure 1: Homepage of ProtParam: Paste the sequence in the given box or enter accession number.
And for this number of amino acids, molecular weight, theoretical PI, amino acid composition, total number of atoms, extinction coefficient, instability index, aliphatic index, grand average of hydropathicity, total number of negatively and positively charged residue and estimated half -life is computed. If instability index is below 40 then the protein is predicted as stable and above 40 it may be unstable, the aliphatic index ascertains the thermal stability based on amino acids alanine, valine and leucine of globular proteins. If Gravy value is low then this deciphers that there is better interaction between protein and water. Half- life predicts the amount of time taken by half of protein to disappear after its production in cell, the extinction coefficient predicts amount of light absorbed by a protein at certain wavelength. If amino acids are basic in nature then PI will be high and if amino acids are acidic then the PI will be low [3].
Secondary structure prediction
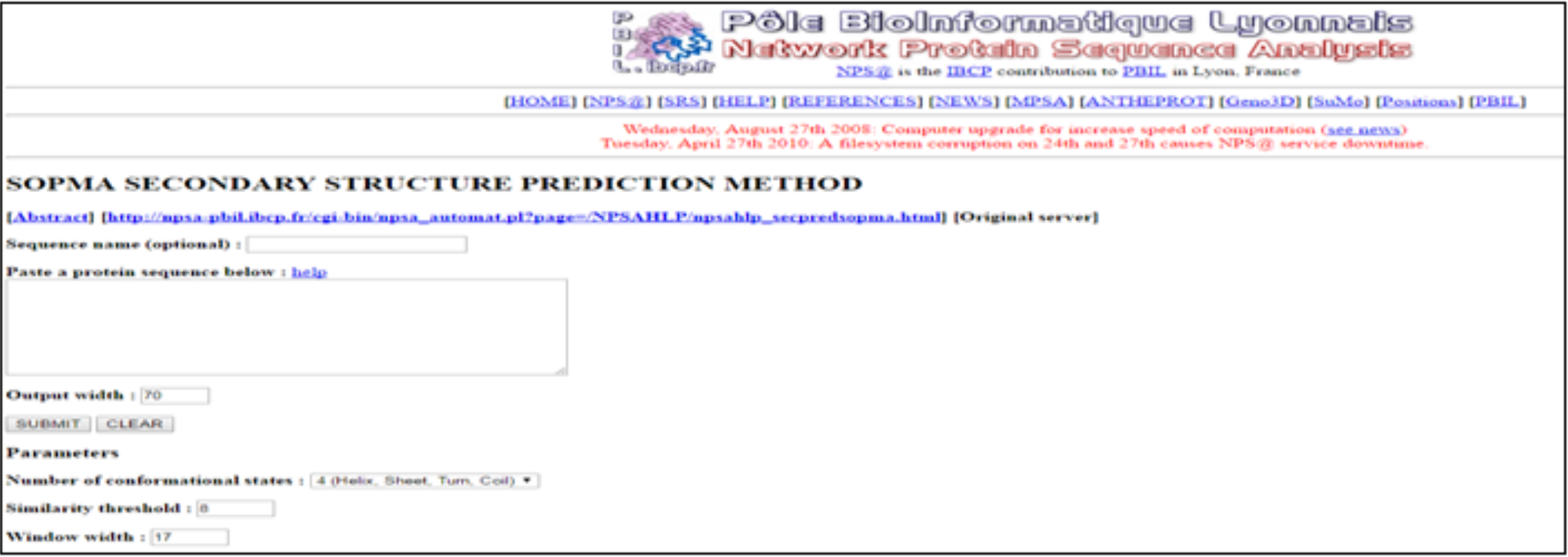
The secondary structure of protein is predicted using SOPMA (Self- Optimized Prediction Method with Alignment) tool (Figure 2).
Figure 2: Homepage of SOPMA: Paste the sequence in the given box and choose parameters accordingly.
This tool evaluates the percentage of alpha helices, extended strand, beta turn and random coils. It uses homology methodology. According to percentage secondary structure is predicted. By default it shows output width as 70 which means there will be 70 amino acids in each line. Number of conformational states can be given as either 4(Helix, Sheet, Turn, Coil) or as 3 (Helix, Sheet, Coil). The first graph of sopma result anticipates the prediction and the second graph consist of outcome curves for all of the predicted states. Other tool that can be used for secondary structure prediction is GOR IV [4].
Tertiary structure prediction
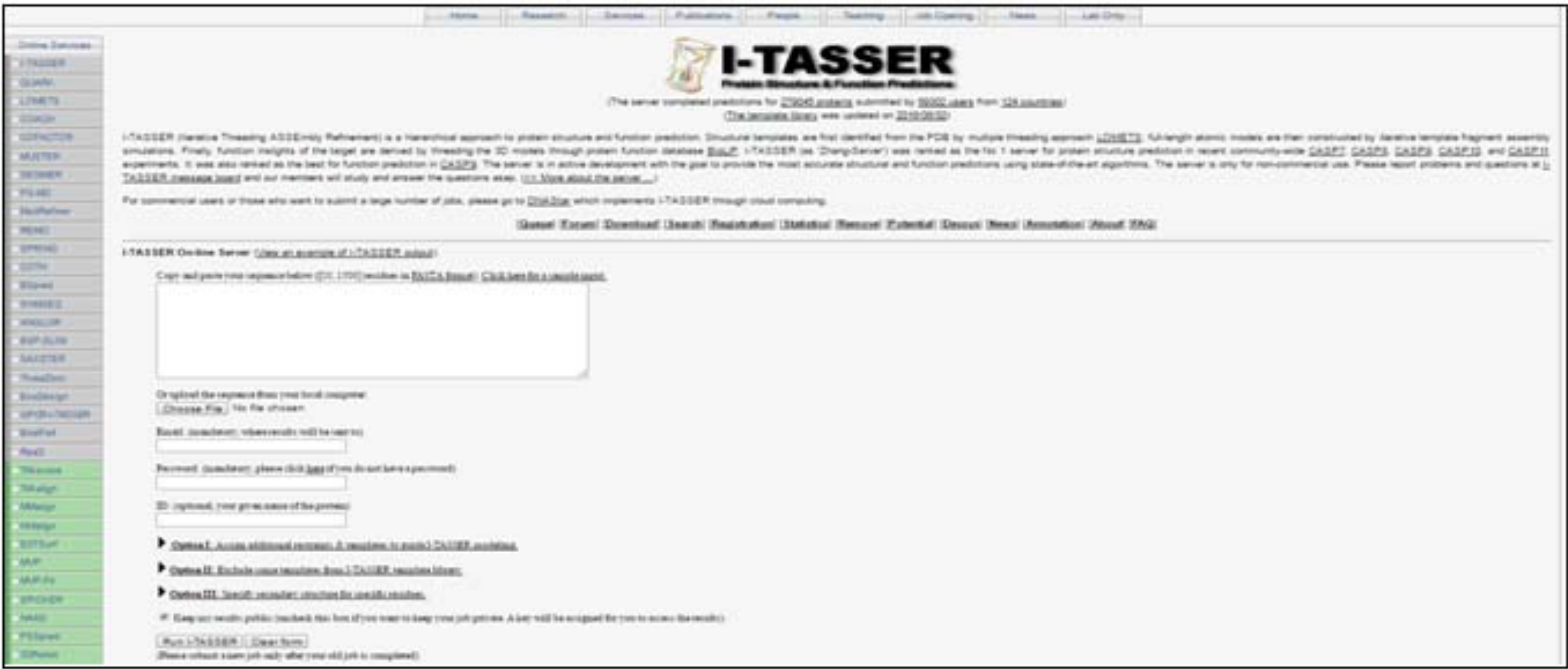
Based on availability of template sequence modeling can be Comparative/Homology or Threading or Ab Initio modeling. I - TASSER (Iterative Threading Assembly Refinement) is one of the best tool for automated protein structure prediction [5]. Solvent Accessibility, Normalized B-factor is predicted [6]. Tools provides top 10 threading templates used by I-TASSER and predicts top (Figure 3)
Figure 3: Homepage of I-TASSER: Paste the sequence in the given box.
ranked 5 models based on C-score, Estimated TM – score and Estimated RMSD. Enzyme commission number and active sites are also predicted [7].
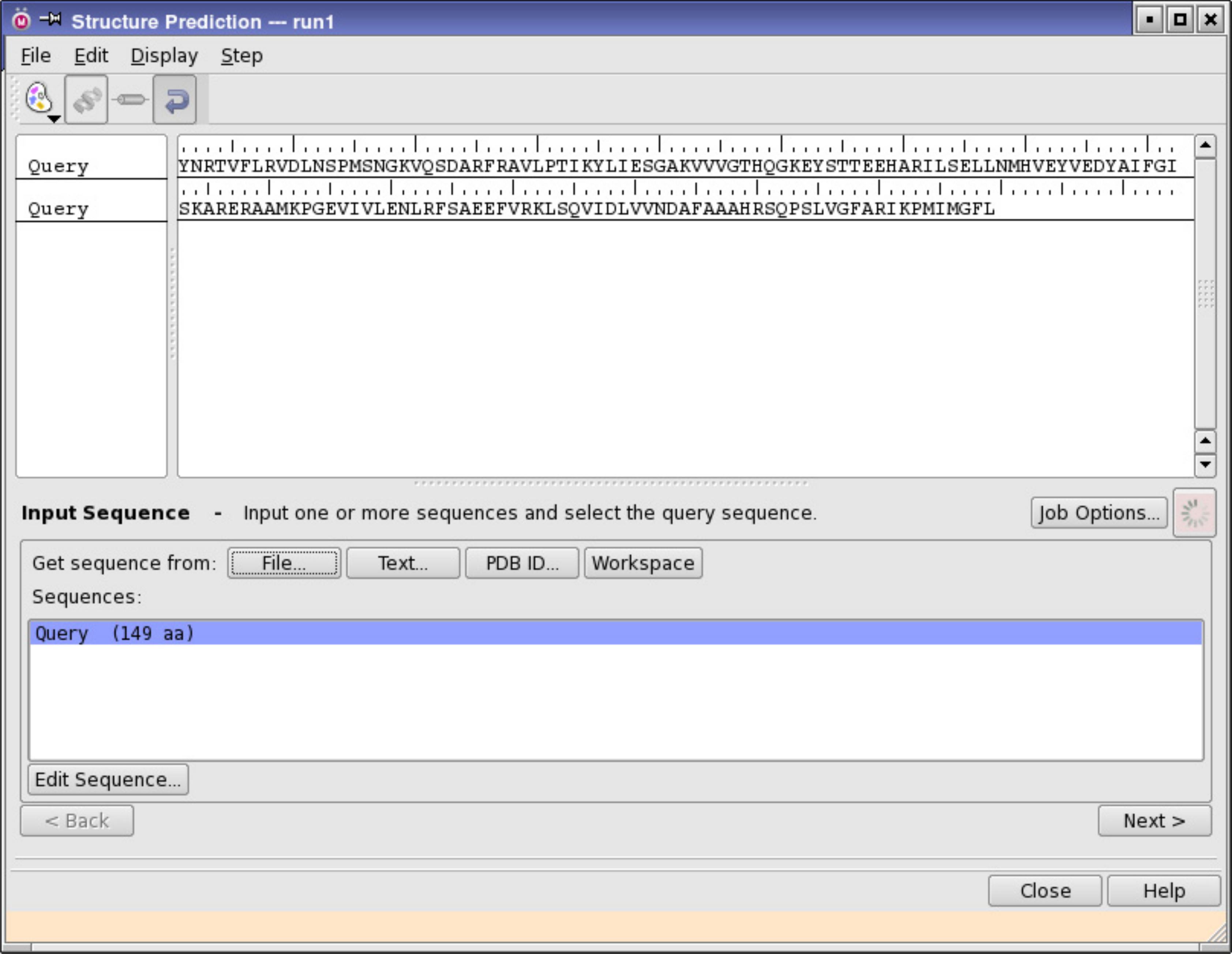
Prime module of Schrödinger LLC, New York, 2014: Taking into account the degree of similarity in sequence protein can be modeled using this tool. Alignment of sequence can be ameliorated manually and homology methodology is utilized to build the structure of protein [8] (Figure 4).
Figure 4: Input sequence in the structure prediction panel.
Other tools used are SWISS-MODEL, Modeller for homology modeling.
Validation of predicted models
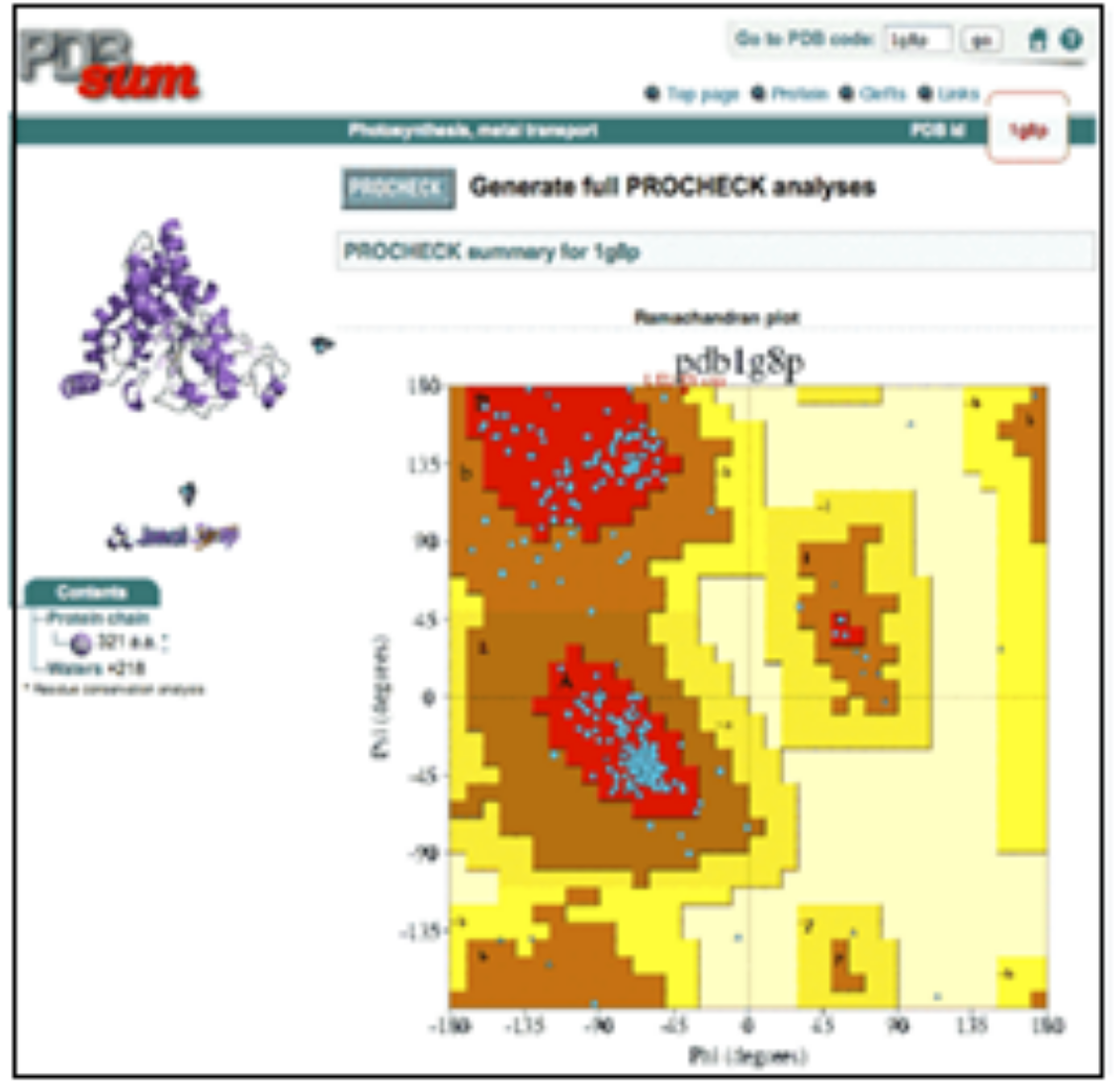
PROCHECK is a tool to substantiate the spatial arrangement (stereochemistry) of protein structures. The output of this program is number of plots which are in PostScript format (Figure 5).
Figure 5: Ramachandran plot of procheck.
The plot generated by this program is Ramachandran plot which is a plot of phi-psi torsional angles where darkest region is the most favoured region consisting of more than 90% of residues. It also generates Ramachandran plot based on types of residues [9].
RAMPAGE is used to check the stereo chemical properties of protein structure whether it is available through experiments or has been modeled (Figure 6).
Figure 6: Homepage of RAMPAGE: Upload PDB file.
The plot generated by RAMPAGE provides the percentage of residues in various regions like favoured region, allowed region and outlier region. The more number of residues in favoured region the more stable is the protein [10].
Phylogenetic Tree
Multiple sequence alignment is an important requirement for additional evaluation of families of protein such as comparative modeling and phylogenetic reestablishment. T-COFFEE (Tree based Consistency Objective Function for alignment Evaluation) is for multiple sequence alignment (Figure 7).
Figure 7: Homepage of T-COFFEE, Paste the sequence in the given box, set parameters if required and submit your job.
T-COFFEE is a program to calculate, manipulate and analyse multiple alignments of RNA (ribonucleic acid), DNA (deoxyribonucleic acid) and protein structures. Minimum of two sequences in supported format is entered as an input or file of sequence can be uploaded [11].
Prediction of Active Site of protein
Pockets present on surface of protein and amino acid residues present in those pockets are substantial for generating physiochemical properties which are required for protein to perform its function. CASTp is an online tool which analyze the active site of the protein and the amino acid that are present in those sites (Figure 8).
Figure 8: Homepage of CASTp.
It provides information of those amino acid residues that would be binding with ligand [12].
‘SiteMap’, Schrödinger LLC, New York, 2014 is also used to predict the active binding site (Figure 9).
Figure 9: SiteMap panel: Select the parameters accordingly and run the job.
Site Score is generated and site maps are ranked. Further these site maps are utilized for generating grid of receptor [13].
Predicting phosphorylation sites
Phosphorylation is a vital procedure through which signaling pathways function. The removal or addition of phosphate group may result in alteration in function of protein and its localization. Three major amino acid residues namely Serine, Threonine and Tyrosine are mostly phosphorylated, as they contain hydroxyl group in their side chain and thus are capable of binding phosphate group. NetPhos server is a tool to predict phosphorylation site at threonine, serine and tyrosine (Figure 10).
Figure 10: Homepage of NetPhos server: Paste the sequence in the given box and submit the job.
The result of NetPhos consists of three parts. Firstly result consists of length and amino acid that are in the sequence. If the amino acid residue is predicted as not phosphorylated (score is below threshold level) then the position is represented as (.) and if it is phosphorylated (score is above threshold level) then residues are marked as ‘T’, ‘S’ and ‘Y’. Prediction for residue is delineated in the second part. And a graph describes the prediction in the third part.
Predicting protein ubiquitination sites
Protein ubiquitination is one of the most vital post-translational modifications by covalent attachment of ubiquitin to lysine residues (Figure 11).
Figure 11: Homepage of mUbiSiDa: Options are selected accordingly.
mUbiSiDa is a universal database for ubiquitination of proteins. The tool provides following functions: advanced retrieval, browse resource and blast search
Prediction of methylation and acetylation
Prediction of potential methylation and acetylation of protein sequence is done using In silico tool PLMLA (Prediction of potential lysine methylation and lysine acetylation) (Figure 12).
Figure 12: Homepage of PLMLA tool. Sequence is pasted in FASTA format.
Sequence is submitted in FASTA format and appropriate option is selected based on post-translational modification requirement for prediction. Name of protein, site position predicted result is returned.
CyMate: It is a tool to perform In silico evaluation of DNA methylation at cytosine site. It is a simple, quick and automated tool. It is a comprehensive tool.
CONCLUSION
The computational or In silico approach that has been highlighted in this review for predicting the structure and function of unknown protein apprehends the efficacy of various tools of bioinformatics. These tools are pre-requisite in predicting structural and functional features thereby facilitating experimental analysis of proteins. The study of proteins made here can be explored and utilized further so that it can be beneficial for therapeutic purposes.