Identification of Small-Sugars as Modulators of NeurotrophinReceptor Interactions
- 1. Department of Morphophysiology, The Royal and Pontificial Major University of Saint Francis Xavier, Bolivia
- 2. Institute of General Organic Chemistry, Department of Bio-organic Chemistry, Spain
- 3. San Francisco Xavier Royal and Pontifical University of Chuquisaca. Domingo Savio Private University, Bolivia
Abstract
Neurotrophins are growth factors involved in the development and maintenance of the nervous system. The activation of these proteins involves the participation of oligosaccharides present in the surface cell membrane or in its receptors (Trk, p75NTR). This paper performs molecular modeling studies of available chemical databases in order to identify potential small-sugars that could serve as modulators of protein/receptor interactions. A library of 1043 compounds with high similarity to the chondroitin sulfate was downloading from PubChem library to be screened against neurotrophin-4/TrkB complex. Identification of small-sugars as allosteric modulators could control the interaction neurotrophin-receptor which is critically involved in angiogenesis process. A structural knowledge of the interactions of neurotrophin/small-sugars/TrkB complex will help to understand the biochemical pathways in which it is involved.
Keywords
Molecular modeling, Neurotrophins, Trk receptor, Virtual screening, Small-sugars
Citation
Camacho V, Bastida A, Zárate SG (2022) Identification of Small-Sugars as Modulators of Neurotrophin-Receptor Interactions. JSM Chem 9(1): 1057.
ABBREVIATIONS
NT: Neurotrophin; NGF: Nerve Growth Factor; BDNF: Brain-Derived Neurotrophic Factor; Trk: Tyrosine Receptor Kinase; p75NTR: p75 Neurotrophin Receptor; DD: Cell Death; CS: Chondroitin Sulfate; GAGs: Glycosaminoglycans; PDB: Protein Data Base; HTVS: High Throughput Virtual Screening; ADMETox: Absorption, Distribution, Metabolism, Excretion-Toxicity; PAINS: Pan Assay Interference Compounds
INTRODUCTION
Neurotrophin has been implicated in a great number of neurological disorders including Alzheimer and Parkinson’s disease [1]. The family of neurotrophins (NTs) is NGF, BDNF, NT-3, NT-4/5, NT-6 and NT-7 which have around 50% sequence homology [2]. Their activities are mediated by interacting with polysaccharides present on the surfaces of cell membranes or glycosylated receptors [3-6]. Those receptors are transmembrane proteins of two kinds; p75NTR receptor which can bond with all neurotrophins with low affinity and tyrosine receptor kinases (Trks) which are each specific for different neurotrophins (TrkA binds NGF, NT-7, TrkB binds BDNF, NT-3, NT-4/5, and TrkC is specific for NT-3) and have high affinity for those proteins [7,8]. NTs and Trks are highly preserved during evolution and have been detected in all vertebrate nervous systems and in some invertebrate so the origin of the NT/Trk signaling is prior to the origin of vertebrates. The simultaneous activation of Trks at the same time is a result of a wide range of cross-interactions between receptors and neurotrophins.
The structural features of the Trk receptors consists of two cysteine cluster (C1, 2), three leucine-rich motif (Leu1, 2, 3), two immunoglobulin regions (IgC1, IgC-2) and one transmembrane domain to transduce signals which connects to the cytoplasmic region where a tyrosine kinase domain is present (Figure 1)
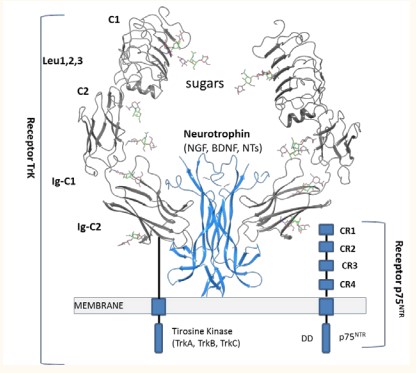
Figure 1: General structure of Neurotrophins (NGF, BDNF, NT-4 or NT-3) bond to the Trk/ p75NTR receptors. Sugars; GlcNAc, GalNAc.
All Trk (TrkA, TrkB and TrkC) receptors are homologous in sequence (amino terminal forming the extracellular portion of the receptor where binding of the neurotrophin takes place). The p75NTR receptor has an extracellular region with cysteine rich regions (CR1-CR4), and one transmembrane domain which connects to the cytoplasmic domain involved in cell death (DD) [9]. The most common polysaccharides found in the extracellular matrix and on cell surfaces are oligosaccharides as heparin or chondroitin sulfate [4,5]. Interaction of neurotrophin with Trk receptors promotes neuron survival, whereas activation of p75NTR gives apoptosis and cell death [9]. Available crystal structures of Trk receptors and neurotrophins are a valuable resource for exploitation via molecular modeling its NT/Trk binding complex which will provide insight to structural regions that may be candidates for drug targeting and signaling pathway selection. NT/Trk interactions depend on many factors such as selectivity, binding affinity, and extracellular or intracellular modulation by other proteins.
In the literature has been described that anionic oligosaccharides as glycosaminoglycans (GAGs) are able to bind neurotrophic factors to form a large complex [10-13]. Chondroitin sulfate (CS) is a relevant family of glycosaminoglycan that participates in a large variety of biological events that are related to neural processes by regulating various GFs. It
polysaccharide is composed of repeating 4)-β-GlcA-(1→3)-βGalNAc-)1→ disaccharide units. CS chains are modified by specific sulfotransferases with a sulfate group at C-2 of the glucuronic acid unit as well as C-4 and/or C-6 of the GalNAc residues, yielding various kinds of monosulfated or disulfated disaccharide units such as [GlcA-4OSGalNAc]n or [GlcA-6OSGalNAc]n or [GlcA-4,6OSGalNAc]n. Distinct modification patterns on CS are the structural basis for their biological functions and activities [14]. CSs have the ability to bind a large number of proteins, in particular to NTs. In the literature it is documented that CS binding site within the NT/Trk complex could be a potential mechanism for how CS promotes complex formation and modulates neurotrophin signaling [3]. Therefore, a structural knowledge of the interactions of neurotrophins with smallsaccharides will help to understand the biochemical pathways in which it is involved. Computational studies are the rational follow-up for structural analysis, as well as prior to functional analysis. So, crystal structure of the receptor and the growth factor is a valuable resource for docking molecular [15-18].
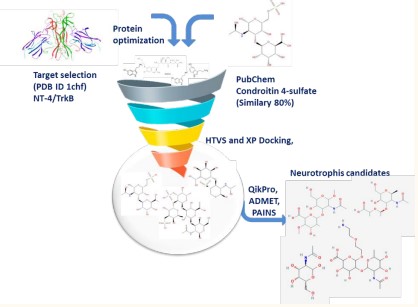
In this study, we have using molecular modeling of available chemical database to identify small-sugars that can be developed into novel neurotophins antagonist. NT-4/TrkB complex were used to perform the molecular modeling of 1043 sugars deposited in the PubChem library that share >80% similarity to the chemical structure of chondroitin sulfated disaccharide.
MATERIALS AND METHODS
he structure file from PDB ID 1hcf of the NT-4/TrkB complex was used to prepare the protein optimization [8] using the Protein Preparation Wizard module integrated within the Maestro suite software (Schrödinger, LLC, New York, NY, USA, 2019). Binding site was detecting using SiteMap from Schrödinger, LLC, New York, NY, 2021.
Chondroitin sulfate (CID 21873177) was used as lead structure to retrieve one library of 1043 compounds with structurally similar (Tanimoto similarity index >0.8) returned from an open chemistry database PubChem [19]. Low-energy 3D conformations of the compounds were retrieved using the Ligan Preparation module within the Maestro suite (Schrödinger, LLC, New York, NY, USA, 2019). Predict pKa values (neutral pH) were employed with the epik software and return all chemically sensible structures using Hammett and Taft methodology [20]. All small-sugars were minimized using the OPLS3e force field implemented in Maestro [21].
Molecular modeling was performed using High Throughput Virtual Screening (HTVS) Glide-dock module integrated in Maestro [22]. The docking grid was built around the amino acids Tyr96, Arg114, Gln94, Tyr329, and Asn350 with setting the dimensions of 20 Å to ensure a range that covered both proteins. Ligand poses generated in that manner were run through a series of hierarchical filters to evaluate ligand interactions with the protein. Upon conformational analyses and structure optimization, the structures contained within each library were analyzed via HTVS using Glide docking as implemented in the Maestro software. Docking glide scores, specific interactions with amino acid in the interface pocket and structural diversity were considered for selecting the most promising ligands. The ADME/Tox parameters were calculated using QikPro integrated in Schrödinger. After that, PAINS (pan-assay interference compounds) identification was obtained using swiss ADME webserver [23] to filter-off molecules containing such chemical groups Scheme 1.
Scheme 1: Workflow for the identification of novel Neutrophin inhibitors
RESULTS AND DISCUSSION
Neurotrophin/Trk complex as a suitable drug target
Neurotrophins (NTs) are a family of homologous proteins synthesized as precursors in a pro-form and secreted in form of mature protein dimers. Only upon dimerization is there a hydrophobic core to stabilize the structure of the growth factor. The binding of neurotrophin to their respective Trk initiates intracellular signals essential for the growth and survival of neurons. Several crystal structures of NTs and neurotrophin/ receptor complexes are deposited in the Protein data Base (PDB) (Table 1).
Table 1: Neurotrophins and neurotrophin/receptor complex deposited in the PDB.
| Neurotrophin | Receptor | PDB | Chain | Organism | Reference |
| DNF/NT-4 | ---- | 1b8m | A, B | H. sapiens | [24] |
| NT-4 | ---- | 1b8k, 1b98 | A, B | H. sapiens | [24] |
| BDNF/NT-3 | ---- | 1bnd | A,B | H. sapiens | [25] |
| NGF - | ---- | 1bet | A,B | Mus muculus | [26] |
| NGF | TrkB | 1www | A,B | H. sapiens | [26] |
| NT-4 | TrkB | 1hcf | A,B,X | H. sapiens | [8] |
| NGF | TrkA/p75 | 2ifj | A-P | H. sapiens | [7] |
| BDNF/NGF | ----- | 1nt3 | A,B | H. sapiens | [2] |
| NGF | TrkA | 1he7 | A,B | H. sapiens | [8] |
proteins (Ser21, Ala19, Cys119, Lys93, Gln94, His299 and Asn350).
3D-structure of neurotrophin/receptor complex reveals common features that may be important in the binding between the neurotrophins with oligosaccharides which would initiate intracellular signals essential for the growth and survival of neurons.
The overall structure of NT’s dimmer interface (homodimer or heterodimer) presents similar patterns of nonpolar contact and hydrogen bonds. The structure of neurotrophin-4 bound to Ig-C2TrkB receptor is showed in Figure 2.
The most commonoligosaccharide ligands for neurothrophins are glycosaminoglicans (Hepara and chondroitin sulfates). So, that process could be control by new small sugars with similarity structural to those oligosaccharides. So, novel small-sugars able to recognize neurotrophins with high affinity could avoid the formation of the NT/Trk complex. In order to identify novel small-sugars as allosteric modulators of neurotrophin/receptor complex a chemical library of compounds that are structurally related to glycosaminoglicans as chondroitin sulfate was carefully screened and analyzed.
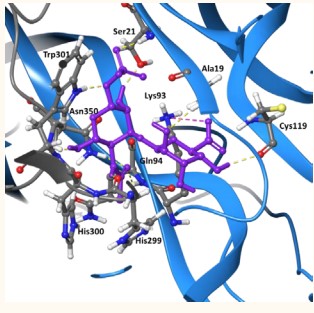
According to the molecular simulation of a ternary structure of the NT/TrkB complex, we determined the amino acid residues that participate in the physical interaction between these two proteins [21]. The identification of binding site of NT-4/TrkB complex was carried out using SiteMap tool from Schrodinger (Figure 2)
Figure 2: 3-D Structure of NT-4 (blue) /Ig-C2TrkB (grey) complex [24] (PDB ID 1hcf).
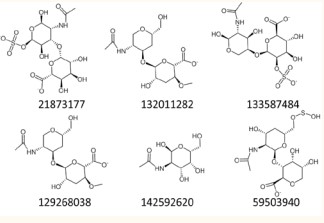
The interface site was employed to define the active site for High Throughput Virtual Screening (HTVS) experiments. A chemical database of compounds sharing >0.8 Tanimoto similarity index with chondroitin sulfate and complying Lipinski’s rules was retrieved from PubChem. This library of 1043 small-sugars was used as the input source for our HTVS upon ligand preparation using the OPLS3 force field. This library of sugars was then subjected to molecular docking studies using the HTVS glide dock module from Schrodinger [19,20]. High precision docking calculations using XP were accomplished for the compounds with the highest docking scores in the HTVS. Hit compound (21873177) and selected candidates based in specific interactions, structural diversity and docking glide scores are shown in Figure 3.
Figure 3: 2D structure of chondroitin sulfate (hit compound, 21873177) and novel small-sugars as modulators of neurotrophin/ TrkB interactions.
Dock of the lead compound within the NT-4/TrkB complex showed structural details of the interactions between neurotrophin-4/receptor complex and chondroitin sulfate (Figure 4)
Figure 4: NT-4(blue)/TrkB(grey) in complex with the lead compound (21873177 stick representation in purple). Interactions are shown in dashed lines; H-bond (yellow); salt bridge (pink).Docking score -9.63 kcal/mol.
(-9.6 kcal/mol docking score). The disaccharide is place in the interface site of both proteins, establishing seven H-bonding and one salt bridge with different residues of both
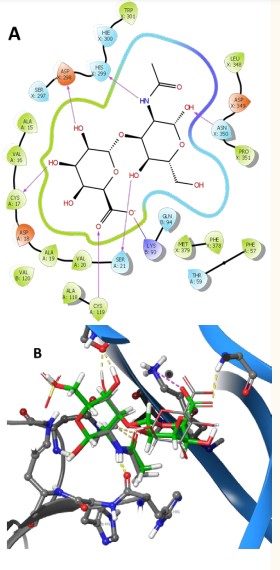
In Figure 5
Figure 5: (A) Ligand interaction of 140843825 with NT/TrkB complex; (B) dock of chondroitin sulfate (grey) and 140843825 (green) in the binding site of complex.
are represented ligand interaction diagrams and dock of one of the best disacharide retrieved from the chemical library (ID 140843825). The novel sugar demostrate that the sulfate part of chondrotin in the GalNAc residue can be substituted by a simple hydroxyl group (-11.33 kcal/mol) or can be present in the GlcA residue (ID 133587484) improving the number of stabilizing interactions with NT-4/TrkB complex.
Ligands 132011282 and 129268038 where hydroxyl groups have been substituted by methyl groups keep the crucial interactions with the protein-receptor complex and even improve the predicted energy of complex formation (Table 2).
| Compound | QPlog | QPlogHERG | QPPCaco | QPlogBB | HOA(%) | Docking score |
| 46836188 | -0.82 | -0.67 | 0.62 | -3.38 | 0 | -12.49 |
| 129268038 | -0.23 | -0.75 | 1.15 | -3.06 | 0 | -11.29 |
| 118118706 | -0.31 | -0.77 | 2.21 | -2.76 | 0 | -10.88 |
| 68320140 | -0.36 | -0.54 | 1.86 | 2.70 | 0 | -10.77 |
| 133587484 | -0.35 | 1.28 | 0.12 | -3.33 | 0 | -10.60 |
| 5159033 | 0.45 | 0.66 | 0.007 | -5.29 | 0 | -10.51 |
| 59679785 | -0.14 | -0.36 | 2.86 | -2.54 | 0 | -10.43 |
| 132011282 | -0.97 | -1.2 | 7.41 | -2.23 | 33.79 | -10.34 |
| 68320140 | -0.38 | -0.56 | 1.43 | -2.82 | 0 | -10.27 |
| 68355982 | -0.48 | -0.7 | 13.86 | -1.89 | 22.06 | -10.03 |
| 21873177 | -0.18 | 0.89 | 0.016 | -4.46 | 0 | -9.63 |
| 57572422 | -0.20 | -0.53 | 4.88 | -2.41 | 0 | -9.56 |
| 59503940 | -1.43 | -0.96 | 2.53 | -2.69 | 0 | -8.54 |
| 142592620 | -0.15 | -1.9 | 43.37 | -1.64 | 41.24 | -8.42 |
| 156351981 | 0.49 | -1.5 | 0.071 | -4-40 | 0 | -7.5 |
| QPlog, predicted aqueous solubility [-6.5/0.5]; QPlogHERG K+ Channel Blockage (log IC50) [concern below -5]; Apparent QPPCaco-2 cell permeability in nm/s [500 excellent]; QPlogBB, predicted log of the brain/blood partition coefficient [-3.0/1.2]; Human Oral Absorption in GI [<25% is poor]. PAINS were 0 for all compounds | ||||||
The new small-sugars should present a strong interaction pattern and docking scores but also a predicted good druggability profile when compared to their reference compound (Table 2). Fourteen compounds were identified as the most promising drug candidates. The most remarkable small sugars from this library are 132011282 and 142592620 (monosaccharide), which showed good interaction energy (six or five H-bonds
with docking score -10.34 or -8.42kcal/mol respectively) and a predicted better absorption and cell permeability than lead compound (Table 2, line green).
ACKNOWLEDGEMENTS
To Dr. Aghata Bastida for her academic and professional support in the preparation of this article, Institute of General Organic Chemistry, Department of Bio-organic Chemistry, Spanish National Research Council (CSIC), Madrid Spain. To
Dr. Sandra Zárate Segóvia, for her support in the elaboration, revision and comments of this work, Chemical Engineering Career, Universidad Mayor Real and Pontificia San Francisco Xavier de Chuquisaca, Sucre-Bolivia.
CONCLUSION
Today, most available NT modulators have a very high molecular weight along with an unfavorable pharmacogenability profile (oligosaccharides such as chondroitin sulfate or heparin sulfate). In silico methods allow much larger targets to be explored than can be accessed experimentally, and can be used to design new ligands. Our computational modeling studies have identified better novel small sugars with optimal drug-like characteristics. In the clinical field it is known that the use of neurotrophins is very limited due to the large number of signals induced by these neurotrophic factors, however, and thanks to the identification of allosteric modulators of the NT-4/TrkB complex, it is concluded that they could be have favorable implications in the treatment of neurodegenerative diseases such as Parkinson’s, Alzheimer’s, amyotrophic sclerosis and others where a dysregulation and decrease of neurotrophins that prevent the development, maintenance and survival of neurons in the central nervous system has been demonstrated.
REFERENCES
25.Robinson RC, Radziejewski C, Stuart DI, Jones EY. April 4, 1995 Accelerated Publications. 1995; 34: 4139–4146.
![Figure 2 3-D Structure of NT-4 (blue) /Ig-C2TrkB (grey) complex [24] (PDB ID 1hcf).](https://www.jscimedcentral.com/public/assets/images/uploads/image-1679730809-1.jpg)