Antibiotic Scuptibility, Biofim-Forming Capacity and Hemolysin and Gelatinase Production By of Enterococus Spp Isolates from Vaginal Microbiota
- 1. Department of Basic Sciences B, University of Tunis El Manar, Tunisia
- 2. Department of Molecular, Cellular Biology & Biotechnological, University of Monastir, Tunisia
- 3. Department of Clinical Biology B, University of Monastir, Tunisia
Abstract
Initially commensal bacteria of the gastrointestinal tract, Enterococcus species are opportunistic pathogens that can colonize the female vaginal tract, particularly in patients with aerobic vaginitis or after receiving antibiotic treatment. Enterococcus spp., are associated with a wide range of illnesses, particularly in patients with altered host microbiota or immunocompromised conditions.c. Here we assessed the antibiotic susceptibility, the biofilm formation ability and the production of hemolysin and gelatinase in enterococcal isolates from vaginal microbiota with phenotypic characterization methods. Our results revealed the prevalence of E. faecalis strains from vaginal samples. 12.9% of E. faecalis were resistant to glycopeptides while E. faecium isolates showed multi-resistant profile to β-lactamine antibiotics, aminoglycosides and macrolides. Biofilm was detected in thirty isolates, of which four were strongly biofilm-producing and thirteen moderately biofilm-producing. Moreover, hemolysin and gelatinase virulence factors were detected in 83.9% and 89.3% of Enterococcus spp strains respectively. In addition to biofilm formation, our results demonstrated that hemolysin and gelatinase production are significant virulence factors in Enterococcus spp isolated from vaginal microbiota.
Keywords
• Enterococcus Spp
• Vaginal
• Microbiota Antibiotic
• Virulence Factors
Citation
Merghni A, Makhlouf A, Ben Saad M, Ben Nasr A (2025) Antibiotic Scuptibility, Biofim-Forming Capacity and Hemolysin and Gelatinase Production By of Enterococus Spp Isolates from Vaginal Microbiota. JSM Microbiology 11(1): 1064.
INTRODUCTION
The gut, genitourinary tract and oral cavity are among the environments with low redox potential where Enterococci thrive [1]. The expression of different enterococcal characteristics eventually leads to virulence in unfavourable conditions [2]. Most of human enterococcal infections are caused by E. faecalis, which is followed by E. faecium among the Enterococcus species [3]. Because of their capacity to acquire and disseminate antibiotic resistance genes, enterococci are becoming significant nosocomial pathogens, with inherently resistant to antibiotics [4]. Increasing virulence may or may not be linked to increasing antibiotic resistance in bacterial cells, as cost-benefit studies have demonstrated that virulence and antibiotic resistance are two entirely distinct characteristics of bacterial cells [5]. More antibiotic resistance was shown by the Enterococcus strains that produced biofilms as opposed to those that did not [6,7].For instance, a key component of the pathogenicity of Enterococcus faecalis isolated from chronic infections is the capability of biofilms production [8]. In addition to biofilm formation, it has been demonstrated that E. faecalis strains that produce hemolysin are virulent in both human and animal infections [9], and are linked to higher infection severity [10]. E. faecalis produces the protease gelatinase, which hydrolyzes a variety of peptides, including collagen, casein, hemoglobin, and gelatin [11]. In endocarditis, enterococcal gelatinase is the main pathogenic mediator [12]. Our finding aims to assess the antibiotic susceptibility, the biofilm formation ability and the presence of hemolysin and gelatinase in enterococcal isolates from vaginal samplings.
MATERIALS AND METHODS
Bacterial strains
Our study was carried out on 31 Enterococcus spp strains isolated from vaginal samplings undertaken in the microbiology laboratory of the Monastir Maternity and Neonatology Center in Tunisia during 2019.
Confirmation of bacterial identification
Bacteria were isolated on a chromogenic culture medium (CHROMagarTM StrepB, France). After incubation of each sample at 37°C for 24 h, the greenish-blue colonies corresponding to the Enterococcus genus (as opposed to the mauve colonies for Streptococcus) were plated on potassium tellurite agar medium (Bio-Rad, France). This medium distinguishes E. faecalis from other enterococci species. E. faecalis is the only species capable of reducing tellurite to tellurium, producing black colonies on this medium. Confirmation of the Identification was based on morphological and biochemical characteristics. Selected colonies were identified using standard microbiological techniques, namely Gram staining and catalase enzyme testing, followed by biochemical confirmation using the VITEK® 2 compact automated system (BioMerieux®, France) based on advanced colometry technology. After identification, Enterococcus spp strains were stored at -20°C in eppendorfs tubes containing Brain Heart Broth (BHI, Biorad) supplemented with 30% of glycerol.
Antibiotic susceptibility testing
Antibiotic susceptibility testing was carried out using a Vitek® 2 Compact automated system (BioMerieux®, France) according to the recommendation of EUCAST [13]. The principle of this method is based on the determination of the minimum inhibitory concentration (MIC) comparing with standard MIC values listed in the database of the apparatus. Thus, from an overnight bacterial culture of each tested strain, a suspension was prepared in NaCl solution (0.45%) and optic density of the bacterial inoculum was adjusted to 0.5 McFarland using a DensiCHEK densitometer. The antibiogram card (AST-P592) was then inoculated with the prepared bacterial suspension and incubated in the automated system at 35-37°C for 24 hours. The MIC result for each antibiotic was finally obtained with an Advanced Expert System (AESTM) software. Results from this test are interpreted using clinical categorization (S: sensitive, I: intermediate and R: resistant). The antibiotics tested are Ampicillin (AMP), Imipenem (PM), Teicoplanin (TEC), Vancomycin (VAN), Gentamicin (GEN), Streptomycin (STR), Erythromycin (ERY), Tigecycline (TGC), Ciprofloxacin (CIP), Linezolid (LZD), Trimethoprim/Sulfamethoxazole (T/S).
Biofilm formation
Enterococcus spp strains were incubated in Brain Heart Broth (BHI, Biorad) at 37°C for 24h. A 1:100 dilution in BHI broth supplemented with 2% glucose (m/v) was then performed for each strain tested. A total volume of 200μl of each dilution was transferred to 96-well U-shaped polystyrene plates (Nunclon; Nunc, Denmark). Each strain was tested three times. Wells containing only BHI broth with 2% sterile glucose were considered as negative controls. The plate was then incubated aerobically for 24 hours at 37°C. The broth from each well was then aspirated and the plate rinsed three times with phosphate-buffered saline (PBS) to remove non-adhered bacteria. The plate was then dried at room temperature. After the drying step, the biofilm formed by the bacteria was stained with 100μl of 1% crystal violet solution (Merck, France), for 20 min. Excess dye was removed by three successive washes, with 300μl of sterile distilled water. The optical density of each well was measured at 570nm using a microplate reader (Automated Multi skanreader, Italy). The ability of strains to form biofilm was classified into three categories [14]:
- “Strongly biofilm-forming”: (OD570≥DOc)
- “Moderately biofilm-forming”: (2ODc≤DO570<4DOc)
- “Weakly biofilm-forming” (ODc≤DO570<2DOc)
- “Non-biofilm-forming”: (OD570 <DOc)
ODc: Optical density control [14].
Gelatinase production
The gelatinase production capacity of isolated bacteria was determined after inoculation of strains onto TSA media (Trypticase Soy Agar; Biorad) supplemented with 0.8% (m/v) gelatin. The presence of a clear halo around the colonies indicates the presence of the desired enzyme. After detecting the presence of this enzyme, we quantified the gelatinase activity. To do this, we sterilely dug 5mm diameter wells in the gelatin agar (0.8%) prepared beforehand. Then, using the 24h cultures of the enterococci strains to be tested, on TSB liquid medium (Trypticase Soy Broth; Biorad), a volume of 50µl was transferred to the wells already prepared on the agar. After incubation at 37°C for 24 hours, we measured the diameter of the halo around each well [15].
Hemolytic potency
Hemolytic activity was tested on blood agar. Each strain was inoculated by circular streaking using a metal loop. After incubation for 24 hours at 37°C, 3 types of hemolysis can be observed:
- Hemolysis α: greenish in colour, resulting from partial lysis of red blood cells.
- β hemolysis: light yellowish colour, resulting from complete red cell lysis.
- Hemolysis γ: no hemolysis and blood agar retain its red color [16].
After detecting the type of hemolysis, we quantitatively estimated this hemolytic activity. Sterile 5mm diameter wells were dug in the blood agar. Then, using the 24h cultures of enterococci on TSB liquid medium, a volume of 50µl was transferred to the wells already prepared on blood agar. After incubation at 37°C for 24h, we measured the diameter of the hemolysis halo around each well.
RESULTS
Confirmation of bacterial species
100% of the isolated strains were gram-positive with cocci form after Gram staining. All of these strains were catalase producer. Confirmation of enterococcal species identification was carried out biochemically using the Vitek® 2 Compact automated system. 28 strains were identified as E. faecalis (90%) and 3 strains of E. faecium (10%) as presented in Table 1.
Table 1: Confirmation of the bacterial species identification
|
Nomber |
Code |
CHROMagarTM StrepB |
Potassium Tellurite |
Gram staining |
Catalase test |
Vitek (strain) |
|
1 |
16325 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
2 |
16382 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
3 |
14635 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
4 |
15971 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
5 |
53945 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
6 |
54011 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
7 |
54113 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
8 |
54758 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
9 |
53977 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
10 |
54091 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
11 |
54085 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
12 |
55630 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
13 |
56234 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
14 |
55929 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
15 |
56170 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
16 |
55386 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
17 |
55507 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
18 |
55680 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
19 |
57060 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
20 |
56727 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
21 |
57327 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
22 |
58620 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
23 |
59137 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
24 |
59987 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
25 |
17133 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
26 |
53964 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
27 |
58560 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
28 |
59197 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecalis |
|
29 |
14502 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecium |
|
30 |
54709 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecium |
|
31 |
54520 |
Blue-green |
Black colonies |
Gram (+) cocci |
+ |
E. faecium |
Antibiotic resistance
profile The results of the resistance profile of enterococcal isolates showed that 4 strains (12.9%) of E. faecalis were resistant to vancomycin and teicoplanin (Glycopeptide family), which are considered a treatment of last resort against these species. In addition, 3 strains of E. faecium (9.67%) are multi-resistant to β-lactam antibiotics, aminoglycosides and macrolides (Table 2).
Table 2: Antibiotic resistance profiles of E. faecalis and E. faecium strains.
|
Strain |
AMP |
PM |
TEC |
VAN |
GEN |
STR |
ERY |
TGC |
CIP |
LZD |
T/S |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
I |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
S |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
S |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
I |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
s |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
S |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
I |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
I |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
I |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
I |
S |
* |
S |
I |
|
E. faecalis |
S |
S |
S |
S |
S |
S |
I |
S |
* |
S |
I |
|
E. faecalis |
I |
S |
R |
R |
S |
S |
R |
S |
* |
S |
I |
|
E. faecalis |
I |
S |
R |
R |
S |
S |
R |
S |
* |
S |
I |
|
E. faecalis |
I |
S |
R |
R |
S |
S |
R |
S |
* |
S |
I |
|
E. faecalis |
I |
S |
R |
R |
S |
S |
R |
S |
* |
S |
I |
|
E. faecium |
R |
R |
S |
S |
R |
R |
R |
S |
R |
S |
I |
|
E. faecium |
R |
R |
S |
S |
R |
R |
R |
S |
R |
S |
I |
|
E. faecium |
R |
R |
S |
S |
R |
R |
R |
S |
R |
S |
I |
Biofilm formation
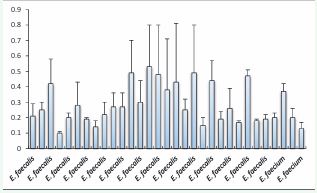
The results of biofilm formation abilities were presented in Figure 1.
Figure 1 Biofilm formation abilities of Enterococcus spp isolates from vaginal microbiota. assed by the crystal violet staining methods. Values are the average of at least three independent determinations.
The Enterococcus spp strains exhibited different level of biofilm formation capability. We have found that 4 out of 28 of E. faecalis strains were highly biofilm-forming (DO570≥0.48), 12/28 strains were moderately biofilm-forming (0.24≤DO570<0.48), 11/28 strains were weakly biofilm-forming (0.12≤DO570<0.24), and only one strain that was unable to form a biofilm, representing 14.3%, 42.9%, 39.3%, and 3.5% of the total tested strains respectively. Regarding E. faecium strains, only one strain tested was moderately biofilm-forming (33.3%). However, the two other strains were found to be weakly biofilm-forming.
Gelatinase production Gelatin hydrolysis was detected by the appearance of a clear halo around the wells on gelatin agar (Figure 2).
Figure 2 Gelatinase (A) and Hemolysin production by tested Enterococcus spp strain. Graph in the right showed the diameter of the well (initially containing the bacterial suspension) and the zone of inhibition (degradation of the substrate) revealing the secretion of the enzyme or toxin by the tested strain.
Our results showed that 100% of E. faecium strains were able to produce gelatinase, compared with E. faecalis (89.3%). Moreover, it was also found that 20 isolated Enterococcus strains presented diameters exceeding 12mm. For instance, the Strain 58620 showed a hemolysis diameter of 14.5mm (Table 3).
Table 3: Biofilm formation, gelatinase and hemolysin production by vaginal Enterococcus spp strains.
|
Strains |
Biofilm formation |
Gelatinase production |
Hemolysin production |
|||
|
|
OD (570) |
Ability |
Qualitative |
Quantitative |
Qualitative |
Quantitative |
|
E. faecalis |
0.21±0.08 |
F |
+ |
14±1.4 |
Hemolysisß |
6.5±0.40 |
|
E. faecalis |
0.25±0.05 |
M |
+ |
12±0 |
Hemolysisß |
8.33±0.47 |
|
E. faecalis |
0.42±0.16 |
M |
+ |
13±1.4 |
Hemolysisß |
15±0 |
|
E. faecalis |
0.10±0.01 |
NF |
+ |
12±0 |
Hemolysisß |
15±0 |
|
E. faecalis |
0.20±0.03 |
F |
- |
5±0 |
Hemolysisß |
13.66±0.94 |
|
E. faecalis |
0.28±0.15 |
M |
+ |
12.5±0.7 |
Hemolysisß |
14.33±0.47 |
|
E. faecalis |
0.19±0.01 |
F |
+ |
12±1.4 |
Hemolysisß |
13.5±0.5 |
|
E. faecalis |
0.14±0.04 |
F |
+ |
13±1.4 |
Hemolysisß |
15±0 |
|
E. faecalis |
0.22±0.08 |
F |
+ |
12±0 |
Hemolysisß |
17±2 |
|
E. faecalis |
0.27±0.09 |
M |
+ |
12±1.4 |
Hemolysisß |
15±1 |
|
E. faecalis |
0.27±0.09 |
M |
+ |
14± |
Hemolysisß |
15.5±0.5 |
|
E. faecalis |
0.49±0.21 |
H |
+ |
11.5±0.7 |
Hemolysisß |
15±1 |
|
E. faecalis |
0.30±0.14 |
M |
+ |
12±1.4 |
Hemolysisß |
14±0 |
|
E. faecalis |
0.53±0.27 |
H |
+ |
11±0 |
Hemolysisß |
15±1 |
|
E. faecalis |
0.48±0.32 |
H |
- |
5±0 |
Hemolysisß |
8±0 |
|
E. faecalis |
0.38±0.33 |
M |
+ |
12.5±0.7 |
Hemolysisß |
7±0 |
|
E. faecalis |
0.43±0.38 |
M |
+ |
10.5±0.7 |
Hemolysisß |
7±0 |
|
E. faecalis |
0.25±0.07 |
M |
+ |
10.5±0.7 |
Hemolysisß |
7±0 |
|
E. faecalis |
0.49±0.31 |
H |
+ |
11.5±0.7 |
Hemolysisß |
14.66±0.47 |
|
E. faecalis |
0.15±0.05 |
F |
+ |
13±0 |
Hemolysisß |
15.66±0.47 |
|
E. faecalis |
0.44±0.13 |
M |
+ |
12.5±0.7 |
Hemolysisß |
13±0 |
|
E. faecalis |
0.19±0.05 |
F |
+ |
14.5±0.7 |
Hemolysisß |
13±0±0.47 |
|
E. faecalis |
0.26±0.13 |
M |
+ |
13.5±0.7 |
Hemolysisß |
8.66±0.47 |
|
E. faecalis |
0.17±0.01 |
F |
+ |
13±0 |
Hemolysisß |
13.33±0.47 |
|
E. faecalis |
0.47±0.04 |
M |
+ |
11±0 |
Hemolysisß |
13.66±0.47 |
|
E. faecalis |
0.18±0.01 |
F |
- |
5±0 |
Hemolysisß |
12.33±0.47 |
|
E. faecalis |
0.19±0.03 |
F |
+ |
12.5±0.7 |
Hemolysisß |
11.33±0.47 |
|
E. faecalis |
0.20±0.03 |
F |
+ |
12.5±0.7 |
Hemolysisß |
11.33±0.47 |
|
E. faecium |
0.37±0.05 |
M |
+ |
10.5±0.7 |
Hemolysisß |
12.66±0.47 |
|
E. faecium |
0.20±0.06 |
F |
+ |
13.5±0.7 |
Hemolysisß |
15.33±0.94 |
|
E. faecium |
0.13±0.04 |
F |
+ |
11.5±0.7 |
Hemolysisß |
15.66±0.47 |
Hemolysin production Firstly, the Hemolysin production was revealed qualitatively by the presence of a clear halo around colonies seeded on blood agar (Figure 2). We noted that 26 strains of enterococci exhibited β-type hemolysis (83.9%) versus 5 γ-type strains (16.1%). Then, the results of the quantitative test showed that 11 strains of E. faecalis had hemolysis diameters exceeding12mm, and up to 17mm (Strain 56234). In contrast, the 3 E. faecium strains had smaller diameters, ranging from 8 to 11.5 mm (Table 3).
DISCUSSION
Enterococci, typically found in intestinal and skin microbiota, are microorganisms with a remarkable capacity to develop resistance to antibiotics [17]. The prevalence of vancomycin resistance in these organisms is rising in hospitals, which limits the available treatments [18]. Despite their relatively modest virulence, they can seriously infect susceptible patients, particularly with impaired immune systems and/or those in need of complex long-term care. Enterococcus faecalis and Enterococcus faecium are the two primary species. In the present study 90.3 % of the isolates were identified as E. faecalis while 9.7% belongs to E. faecium species. This is in agreement with previous reports showing that the prevalence of E. faecalis strains from vaginal origin compared to E. faecium [7,19]. The antibiotic susceptibility test from our study revealed that 4 strains of E. faecalis were resistant to vancomycin and teicoplanin (12.9%) which belong to Glycopeptide family, considered as a treatment of last resort against this species. Although the epidermis, oropharynx and gastrointestinal tract are the most common sites of colonization by vancomycin-resistant enterococci [20], our results underlined that the vagina is another possible niche for the colonization with this bacterial phenotype. More generally, the clinical effectiveness of vancomycin is at risk due to the emergence and widespread dissemination of two types of complicated resistance mechanisms, each consisting of a multi-enzyme route, in pathogenic species [21]. Additionally, the tested strains of E. faecium showed multi-resistant to β-lactam aminoglycosides and macrolides antibiotics. It was reported that unlike E. faecalis, E. faecium is more frequently drug-resistant [22]. In line with our research, several studies on E. faecium strains show that these strains are very sensitive to tigecycline, highlighting their potential as suitable therapeutic options for treating resistant infections caused by this bacterium [23, 24]. Because of its propensity to build biofilm highly than E. faecium, the treating of E. faecalis infections is particularly difficult and may be a factor in the stagnant mortality rates [25,26]. This is an agreement with our finding since we have found that the majority of E. faecalis isolates were found to be biofilm forming strain. A recent report showed that 68.27% of E. faecalis strains, collected from different sources environmental and clinical samples, were biofilm producers [27]. More generally, biofilm-associated illnesses are challenging to cure, because inside biofilms bacteria are resistant to phagocytosis, antibiotics and environmental stress [28,29]. Hemolysin, aggregation substance, and gelatinase have all been identified as enterococci’s virulence factors [15]. Our finding revealed that more than 89.3% of clinical strains of Enterococcus spp isolated from vagina were gelatinase producer. We also noted that 83.9% od the tested strains exhibited β-hemolysis. It was previously showed that 64% blood isolates of E. faecalis from patients with bacteremia were gelatinase producer [9]. Additionally, increased infection severity has been linked to hemolysin producing strains. This cytolytic protein has the ability to lyse human erythrocytes causing cell damages [30].
CONCLUSION
The identification of virulence factors linked to enterococcal infections in women will be a crucial focus of future research due to the growing significance of Enterococcus species as nosocomial pathogens and the rising incidence of glycopeptide resistance among enterococci. In the context of antibiotic resistance, the search for new molecules with preventing or suppressing properties against enterococcal virulence factors could offer therapeutic alternatives for combating these pathogenic bacteria.
REFERENCES
- Lebreton F, Willems RJL, Gilmore MS. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In: Gilmore MS, Clewell DB, Ike Y, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston: Massachusetts Eye and Ear Infirmary. 2014.
- Geraldes C, Tavares L, Gil S, Oliveira M. Enterococcus Virulence and Resistant Traits Associated with Its Permanence in the Hospital Environment. Antibiotics (Basel). 2022; 11: 857.
- Wang Y, Xu W, Guo S, Xu S, Wang J, Zhang S, et al. Enterococci for human health: A friend or foe? Microb Pathog. 2025; 201: 107381.
- Abdel-Raheem SM, Khodier SM, Almathen F, Hanafy AT, Abbas SM, Al-Shami SA, et al. Dissemination, virulence characteristic, antibiotic resistance determinants of emerging linezolid and vancomycin- resistant Enterococcus spp. in fish and crustacean. Int J Food Microbiol. 2024; 418: 110711.
- Schroeder M, Brooks BD, Brooks AE. The Complex Relationship between Virulence and Antibiotic Resistance. Genes (Basel). 2017; 8: 39.
- Sie?ko A, Wieczorek P, Majewski P, Ojdana D, Wieczorek A, Olsza?ska D, et al. Comparison of antibiotic resistance and virulence between biofilm-producing and non-producing clinical isolates of Enterococcus faecium. Acta Biochim Pol. 2015; 62: 859-866
- Sengupta M, Sarkar S, SenGupta M, Ghosh S, Sarkar R, Banerjee P. Biofilm Producing Enterococcus Isolates from Vaginal Microbiota. Antibiotics (Basel). 2021; 10: 1082.
- Lee K, Lee K-M, Kim D, Yoon SS. Molecular Determinants of the Thickened Matrix in a Dual-Species Pseudomonas aeruginosa and Enterococcus faecalis Biofilm. Appl. Environ Microbiol. 2017; 83: e01182-17.
- Vergis EN, Shankar N, Chow JW, Hayden MK, Snydman DR, Zervos MJ, et al. Association between the presence of enterococcal virulence factors gelatinase, hemolysin, and enterococcal surface protein and mortality among patients with bacteremia due to Enterococcus faecalis. Clin Infect Dis. 2002; 35: 570-5.
- Giridhara Upadhyaya PM, Umapathy BL, Ravikumar KL. Comparative study for the presence of enterococcal virulence factors gelatinase, hemolysin and biofilm among clinical and commensal isolates of Enterococcus faecalis. J Lab Physicians. 2010; 2: 100-4.
- Giridhara Upadhyaya PM, Ravikumar KL, Umapathy BL. Review of virulence factors of enterococcus: an emerging nosocomial pathogen. Indian J Med Microbiol. 2009; 27: 301-305.
- Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun. 2010; 78: 4936-43.
- EUCAST: European Committee on Antimicrobial Susceptibility Testing – EUCAST. 1997.
- Haddad O, Merghni A, Elargoubi A, Rhim H, Kadri Y, Mastouri M. Comparative study of virulence factors among methicillin resistant Staphylococcus aureus clinical isolates. BMC Infect Dis. 2018; 18: 560.
- Kanemitsu K, Nishino T, Kunishima H, Okamura N, Takemura H, Yamamoto H, et al. Quantitative determination of gelatinase activity among enterococci. J Microbiol Methods. 2001; 47: 11-16.
- R?ži?ková M, Vít?zová M, Kushkevych I. The characterization of Enterococcus genus: resistance mechanisms and inflammatory bowel disease. Open Med. 2020; 15: 211-224.
- Farsi S, Salama I, Escalante-Alderete E, Cervantes J. Multidrug- Resistant Enterococcal Infection in Surgical Patients, What Surgeons Need to Know. Microorganisms. 2023; 11: 238.
- Cimen C, Berends MS, Bathoorn E, Lokate M, Voss A, Friedrich AW, et al. Vancomycin-resistant enterococci (VRE) in hospital settings across European borders: a scoping review comparing the epidemiology in the Netherlands and Germany. Antimicrob Resist Infect Control. 2023; 12: 78.
- Serretiello E, Santella B, Folliero V, Iervolino D, Santoro E, Manente R, et al. Prevalence and Antibiotic Resistance Profile of Bacterial Pathogens in Aerobic Vaginitis: A Retrospective Study in Italy. Antibiotics (Basel). 2021; 10: 1133.
- Zirakzadeh A, Patel R. Vancomycin-resistant enterococci: Colonization, infection, detection, and treatment. Mayo Clin Proc. 2006; 81: 529-536.
- Stogios PJ, Savchenko A. Molecular mechanisms of vancomycin resistance. Protein Sci. 2020; 29: 654-669.
- Baddour LM, Wilson WR, Bayer AS, Fowler VG Jr, Tleyjeh IM, Rybak MJ, et al. American Heart Association Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and Stroke Council. Infective Endocarditis in Adults: Diagnosis, Antimicrobial Therapy, and Management of Complications: A Scientific Statement for Healthcare Professionals From the American Heart Association. Circulation. 2015; 132: 1435-86.
- Dadashi M, Sharifian P, Bostanshirin N, Hajikhani B, Bostanghadiri N, Khosravi-Dehaghi N, et al. The global prevalence of daptomycin, tigecycline, and linezolid-resistant Enterococcus faecalis andEnterococcus faecium strains from human clinical samples: a systematic review and meta-analysis. Front Med. 2021; 8: 720647.
- Guan L, Beig M, Wang L, Navidifar T, Moradi S, Tabaei FM, et al. Global status of antimicrobial resistance in clinical Enterococcus faecalis isolates: systematic review and meta-analysis. Ann Clin Microbiol Antimicrob. 2024; 23: 80.
- Di Rosa R, Creti R, Venditti M, D’Amelio R, Arciola CR, Montanaro L, et al. Relationship between biofilm formation, the enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol Lett. 2006; 256: 145-150.
- Conwell M, Dooley JSG, Naughton PJ. Enterococcal biofilm-A nidus for antibiotic resistance transfer? J Appl Microbiol. 2022; 132: 3444- 3460.
- Ullah MA, Islam MS, Rana ML, Ferdous FB, Neloy FH, Firdous Z, et al.Resistance Profiles and Virulence Determinants in Biofilm-Forming Enterococcus faecium Isolated from Raw Seafood in Bangladesh. Pathogens. 2023; 12: 1101.
- Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010; 35: 322-332.
- Ghazvinian M, Asgharzadeh Marghmalek S, Gholami M, Amir Gholami S, Amiri E, Goli HR. Antimicrobial resistance patterns, virulence genes, and biofilm formation in enterococci strains collected from different sources. BMC Infect Dis. 2024; 24: 274.
- Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, Clewell DB, et al. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993; 37: 2474-2477.