Prognostic Evaluation of a Urine-Based Methylation Biomarker in Cell-Free DNA for Early Detection of Prostate Cancer
- 1. Department of Cell Stress Biology, Roswell Park Comprehensive Cancer Center, USA
- 2. Department of Surgery, WNY Healthcare System, USA
- 3. Department of Biostatistics and Bioinformatics, Roswell Park Comprehensive Cancer Center, USA
- 4. Department of Urology, Western New York Urology Associates Cheektowaga, USA
- 5. Department of Cancer Genetics & Genomics, Roswell Park Comprehensive Cancer Center, USA
Abstract
Prostate Cancer (PCa) remains one of the most diagnosed malignancies among men, with current diagnostic methods presenting significant limitations. The Prostate-Specific Antigen (PSA) test, while widely used, lacks specificity and can yield elevated results due to benign factors; some of these factors include physical activity, sexual activity, or medication use which can often lead to unnecessary and invasive biopsies. The Digital Rectal Exam (DRE), another diagnostic tool, is underutilized due to patient discomfort and stigma, particularly among African American (AA) men. This hesitancy contributes to a disproportionate burden of advanced-stage PCa diagnoses and poorer outcomes in the AA community, who are typically diagnosed 3 to 5 years earlier and with more aggressive disease compared to Non-Hispanic White (NHW) men.
To address these disparities and improve diagnostic accuracy, this study explores the potential of urinary biomarkers, specifically utilizing cell-free DNA (cfDNA), as a non-invasive screening tool. Cell-free DNA, released by prostate cancer cells into prostatic fluid and subsequently into urine, has shown promise in correlating with disease stage and clinical progression. The development of a urine-based cfDNA screening assay, coupled with Next Generation Sequencing (NGS) to identify gene signatures, offers a novel approach to stratify patients by risk and guide treatment decisions. This method is particularly advantageous for populations reluctant to undergo traditional screening and is well-suited for deployment in community health settings. The findings support the advancement of cfDNA as a reliable, accessible, and equitable alternative to current PCa screening modalities.
Keywords: Urine; Cell Free DNA; Screening and Prostate Cancer; Prostate-Specific Antigen
Abbreviations: PCa: Prostate Cancer; AA: African American; PSA: Prostate-Specific Antigen; DRE: Digital Rectal Exam; NHW: Non-Hispanic White; BPH: Benign Prostate Hyperplasia; cfDNA: Cell Free DNA
Introduction
Furthermore, African American (AA) men are more likely to be diagnosed with more aggressive and later stages of PCa [2,3]. Additionally, the rate of prostate cancer diagnosis for AA men has been increasing year over year for nearly a decade [4]. This points to a breakdown of early screening for PCa within these communities. This is most commonly due to limited access to health care, socioeconomic status, and the lack of participation in early detection programs [3,5]. More specifically, knowledge, social disparity, treatment options, health inequality, and environmental factors may contribute to why AA men develop PCa more impulsively than Non-Hispanic White (NHW) men in the United States.
In fact, race is considered to be both a risk factor and a prognostic factor with PCa in addition to decreased early detection. The lifetime risk of developing PCa and possibly dying from the disease is substantially higher among AA men than among NHW men [2-4]. Multiple factors (clinical, socioeconomic, and pathologic) have been shown to account for 15% of the increased risk of prostate cancer diagnosis in AA men [6]. However, even though AA exhibit higher risk than NHW men for developing PCa, it has been shown that AA men are also less likely to seek and utilize early detection for PCa. This can be attributed to both structural disparities, such as lack of detection programs and economic barriers to screening events, and sociological factors. Some of these factors may include lack of education and knowledge about PCa, lack of family history which indicates risk, and cultural barriers surrounding getting treatment [5,7]. These health disparities can be reduced by hosting community health events where screening is offered [8]. However, turnout to these events is severely limited in part due to the community’s hesitancy over current screening methods, mainly it is well published that many men do not like the DRE [5,9,10]. These problems with the PSA test and the DRE are particularly problematic because early detection is the best method for detecting PCa, catching the disease before it progresses to the fatal late stages of disease.
While race and age are major risk factors for PCa, there are many other factors which contribute to an individual’s risk and severity of PCa. Veterans and servicemembers have been shown to be at significantly higher risk for PCa. Indeed, servicemembers have between 6 and 8 times the rate of diagnosis and are diagnosed at a much earlier age compared to civilians [11]. The risk to servicemembers is increased for a multitude of reasons, mainly stemming from high exposure to a variety of carcinogens found within their line of work [12,13]. Additionally, race has a significant impact on the incidence of PCa within the military [14].
Over the past couple of decades, the majority of PCa screening has been accomplished by utilizing the PSA test. This test is pervasive because it can be easily prescribed by a patient’s primary care physician along with other routine bloodwork. While the PSA test has many flaws, elevated serum PSA has been linked to several urological issues. Therefore, after receiving a high PSA test, the next step a patient takes is typically a biopsy. The biopsy will be able to identify the problem directly, but has its specific drawbacks, such as urinary problems and long-term pain associated with the procedure. One major issue with the PSA test is that the patient’s serum level is highly reliant on daily circumstances, such as level of physical and sexual activity [15]. The PSA test also cannot distinguish between cancer and other issues affecting the prostate such as Benign Prostate Hyperplasia (BPH) or prostatitis. This lack of specificity leads to many unnecessary biopsies [16]. Despite the extensive use of PSA screening to assist in early detection, PCa remains one of the leading causes of mortality of men in the United States.
There is evidence that there is a need for a new screening tool to be developed which can accurately predict PCa. Science has progressed in the two decades since the discovery of PSA and more sophisticated biomarkers are available to be tested in a variety of forms. The ideal test for cancer puts convenience for the patient at the forefront, therefore relying on a blood sample is no longer ideal. Urine has become the ideal biospecimen for urological cancer due to a variety of reasons [17]. Urine contains a wide range of potential biomarkers: from macromolecules like genes and proteins shed from a tumor to subtle metabolomics changes influenced by cancer growth. A particular interest is in genetic biomarkers that can be found in the urine that cannot only indicate potential for cancer but can also suggest risk of aggression. Many different labs have already shown the wide variety of DNA markers that can be isolated from the urine which are indicators of PCa. For example, it is well reported that the gene fusion between ERG and TMPRSS2 can be found in the urine of PCa patients and can be a clinical indicator of metastatic disease [18]. In fact, several commercial products have already been created utilizing urine for PCa patients. Mi Prostate Score was developed utilizing this gene fusion along with other genetic markers like PCA3. Another urine test for prostate cancer is Select MDX, which screens urine for the genetic biomarkers DLX1 and HOXC6 along with other clinical risk factors. Recently, the urine test ExoDX Prostate IntelliScore received FDA breakthrough designation allowing for further use in the clinic. ExoDX is a urine biomarker test that looks at the presence of exosome for PCa diagnosis [19]. While ExoDX has a high negative predictive value, it is out matched in diagnostic accuracy, specificity, and selectivity by the other urinary tests discussed here [20]. However, ExoDX has the advantage of not requiring prostate manipulation like SelectMDX and Mi Prostate score. The ideal test would have high diagnostic capability without the need for manipulation of the prostate. To move to the next step will be accomplished by using next generation sequencing of methylated genes. Methylation of genes is an epigenetic control mechanism where genes can be turned active or inactive by the cell through modification rather than mutations [21]. Some genes that can be used as diagnostics markers are listed in Table 1.
There is a need in the community to develop a more robust screening tool, and this test should be developed with the needs of all men in mind. It has been noted that finding and treating PCa at an early stage, leads to a better chance for cure. Therefore, it is recommended that NHW men seek screening at the age of 50 and that AA men start at the age of 40 [22]. Typically, this means PSA screening or DRE. However, as stated above, the PSA test can miss many problematic cancers and can account for unnecessary biopsies. There is a clear and present need for a modern PCa screening tool that can indicate the difference between aggressive disease and benign growth. This test should be accessible to both civilians and servicemembers in the community to identify PCa early.
Materials and Methods
Collection of urine
Participants of this study voluntarily donated urine samples after giving informed consent according to the Internal Review Board (IRB) approved protocol (I44117). The participants were recruited either from the Roswell Park Urology Clinic or from Prostate Cancer Screening Events, such as Cruising for a Cure or the Buffalo Sabers Prostate Cancer Early Detection Event. All patients who are diagnosed with Prostate cancer but who did not have a prostatectomy, or a secondary cancer were admissible to the study. This cohort includes 66 Prostate Cancer patients, and 18 Healthy volunteers (Table 2). Table 3 breaks down the cohort by Gleason score, PSA, and cfDNA yields. Collected Urine was Preserved using Norgen Biotech Urine Collection and Preservation Tubes (cat #18113). The Urine was stored at room temperature. Urine was processed no more than 2 days after collection.
Mag Max cfDNA extraction
Cell-free DNA (cfDNA) was extracted from the urine samples utilizing Applied Biosystems MagMAX Cell-Free DNA isolation kit (A29319) following the suggested protocol (Figure 1). Briefly, samples were first centrifuged at 16,000 x g for 10 minutes at 4°C to precipitate any cell debris. The urine was mixed with a lyse and binding solution which contained the MagMax Cell-Free DNA Magnetic Beads. After vortexing the samples using an Eppendorf mixmate for 10 minutes, the magnetic beads were collected using a Dynamag magnetic. Samples were washed with provided wash solution, then 80% ethanol. After the second ethanol wash, the cfDNA was removed from the magnetic beads with a 0.1X TAE solution then rebound to new magnetic beads to increase purity. Once the cfDNA was rebound to the new set of magnetic beads, the samples were washed once with the provided wash solution then two rounds of 80% ethanol washes. The cfDNA was then collected utilizing 15µl of the provided elution solution. Purified cfDNA were analyzed for purity using both Qubit 4 Fluorometer and Agilent TapeStation.
Table 1
Table 1: Potential Methylated Genes for Prostate Cancer Diagnosis
| Gene | Citation |
| GSTP1 | 24,25,26 |
| APC | 26,27,28 |
| RASSF1 | 26,27,29 |
| TMPRSS2 | 30 |
| CRIP3 | 31 |
| HOXD8 | 31 |
| SFRP2 | 26,32 |
| SPDEF | 33,34 |
| ARHGAP21 | 33 |
| TMEM25 | 34 |
| BRCA1 | 35 |
Table 2
Table 2: Demographic breakdown of Cohort
Table 3
Table 3: Cohort Breakdown by Gleason score relating cfDNA yield
| Normal | Gleason 6 (3+3) | Gleason 7 (3+4) | Gleason 7 (4+3) | Gleason 9 (4+5) | |
| Count | 18 | 19 | 36 | 9 | 2 |
| PSA range | 0.2-18.4 | 2.26-11.36 | 1.77-31.55 | 0.39-19.59 | 9.90 - 11.25 |
| Average | 45% | 76% | 69% | 62% | 65% (63-67) |
| % cfDNA | (14-85) | (38-93) | (8-95) | (33-83) | |
| Yield (ng/µl) | 0.4 (0.1-1.9) | 1.8 (0.2-13.7) | 2.0 (0.2-9.36) | 37.4 | 26 |
| (0.8-97.4) | (2.94-49) |
Abbreviations: PSA: Prostate-Specific Antigen; cfDNA: cell Free DNA;
Table 4
Table 4: Sample yields after protocol optimization.
| Total samples after Optimization | 51 |
| # samples > 10 ng | 48 |
| # samples > 50% cfDNA | 45 |
| Max ng/µl | 97.4 ng/µl |
| Average ng/µl | 5.57 ng/µl |
| Max yield (ng) | 96.6 ng |
| Average yield (ng) | 72.0 ng |
Abbreviations: cfDNA: cell Free DNA
Results
Optimized Extraction of cfDNA from Urine
The goal of harvesting cfDNA from urine is to generate high quality samples for various downstream Next Generation Sequencing (NGS) applications. To perform these studies, samples should be greater than 50% cfDNA and have a total concentration greater than 10 ng. After the method was optimized, variability diminished, and a higher proportion of samples fell within the parameters for NGS. This improved cost efficiency of the study and reduced the need for multiple extraction cycles and purification of samples to remove the genomic DNA contamination. Additionally, many of these samples were of high enough concentration to enable multiple studies and applications (Table 4).
Concentration of cfDNA Provides Insight on Disease Progression.
Before committing samples to NGS, the quantity of cfDNA can key in certain information about the patient. By comparing concentration of cfDNA collected (ng/µl) and grouping samples by Gleason score, a trend develops demonstrating a potential link between quantity of cfDNA and severity. Normal samples have significantly lower cfDNA concentration compared to Prostate Cancer patients. Additionally, tumors containing predominantly a Gleason score of 3 tend to have lower cfDNA than tumors containing higher grade groups (Figure 2).
Interestingly, concentration of cfDNA also tends to increase as the need for definitive therapy increases. Patients currently under active surveillance have a significantly
lower concentration of cfDNA isolated from the urine. Biochemical progression of the disease correlates to increase in cfDNA isolation (Figure 3).
Figure 1
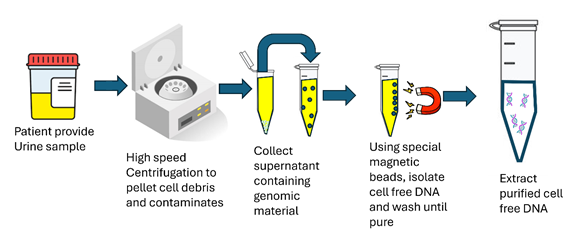
Figure 1: Schematic of cfDNA extraction method from urine.
Figure 2
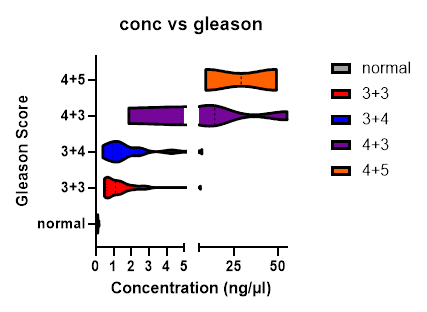
Figure 2: Concentration of cfDNA by Grade Group. Samples are greater than 50% cfDNA and a total yield greater than 10 ng, n=65. 45 samples are from the optimized samples population and 20 samples are of high enough quality before protocol was optimized. Multiple t-tests were performed comparing groups to normal. Gleason 6 (3+3) vs normal p<0.02, Gleason 7 (3+4) vs normal p<0.008, Gleason 7(4+3) vs normal p< 0.03, Gleason 9 (4+5) vs normal p<0.01.
Figure 3
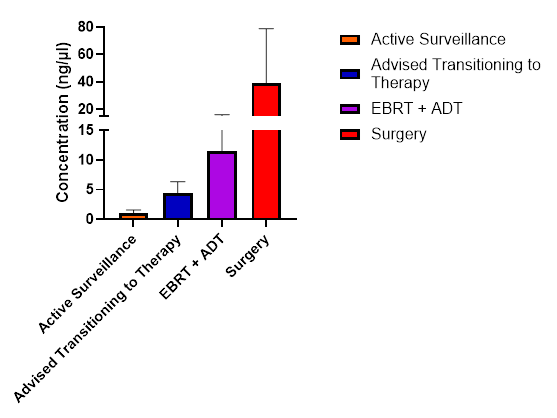
Figure 3: Concentration of cfDNA grouped by patient’s current therapy. Samples under the advised transition to therapy are individuals who are either waiting radiology results to determine best course of treatment or have declined transition to therapy for personal reasons. Multiple t-tests were performed comparing groups to active surveillance. Advised transitioning to therapy vs active surveillance p<0.0001, EBRT+ADT vs active surveillance p<0.0001, Surgery vs Active Surveillance p<0.0001
Discussion
The selection of an appropriate methylation biomarker is critically dependent on the specific clinical context. In this study, we provide foundational insights to support informed decision-making regarding biomarker use and highlight opportunities for further data collection. In the realm of early detection, future research should prioritize rigorous evaluations of test performance across racially diverse populations and explore how these biomarkers can be effectively integrated with Magnetic Resonance Imaging (MRI) and other diagnostic modalities.
Patients with negative initial prostate biopsy results present a significant clinical challenge [23]. Current diagnostic limitations hinder our ability to distinguish between those at high risk of harboring undetected cancer who may benefit from immediate repeat biopsy and those at low risk, who could be spared the morbidity, anxiety, and cost associated with unnecessary procedures. This uncertainty often leads to repeated biopsies, contributing to patient distress and healthcare burden. In this context, the development of a reliable, non-invasive test to complement existing screening and diagnostic tools would represent a substantial advancement in clinical care.
It is also conceivable that a qualitative or quantitative assessment of cfDNA in urine correlate with tumor aggressiveness or provide greater insight on when active treatment should be pursued. If validated, such measures might serve as prognostic indicators akin to the Gleason grade. While this remains speculative, it underscores the potential value of urine-based diagnostics. Our current study provides a compelling rationale for advancing methods that detect prostate cancer in urine with the same rigor and diagnostic standards employed by pathologists in tissue-based assessments.
Conclusion
PCa remains a leading cause of cancer-related morbidity and mortality among men, with AA men disproportionately affected by earlier onset and more aggressive disease. Current diagnostic tools, including the PSA test and DRE, are limited by low specificity, invasiveness, and patient hesitancy particularly within minority populations. These limitations contribute to delayed diagnoses and poorer outcomes in underserved communities.
Emerging research highlights the promise of urinary biomarkers, specifically cfDNA, as a non-invasive and more equitable alternative for PCa screening. Cell-free DNA, shed by prostate cancer cells into prostatic fluid and subsequently into urine, correlates with disease stage and progression. This study explores the potential of cfDNA quantification and gene profiling via NGS to stratify patients by risk and guide clinical decision-making.
Urine-based screening offers a transformative opportunity to overcome intrinsic healthcare barriers, enabling broader participation in early detection efforts especially in community-based and minority-focused settings. The integration of urinary biomarkers into routine clinical practice could revolutionize PCa management by providing a more accurate, accessible, and culturally acceptable diagnostic pathway. Continued innovation in biomarker discovery and validation is essential to realizing this vision and addressing longstanding disparities in prostate cancer care.
Acknowledgements
This work was supported by National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Comprehensive Cancer Center’s Genomic Shared Resource.








































































































































