Surveillance and Genetic Characterization of HPAI H5N1 in Poultry: Evidence from New Valley Governorate, Egypt
- 1. Department of Avian and Rabbit Medicine, New Valley University, Egypt
- 2. Department of Avian and Rabbit Medicine, Assiut University, Egypt
- 3. Department of Aquatic Animal Medicine, New Valley University, Egypt
Abstract
The highly pathogenic avian influenza (HPAI)-H5N1 virus is a highly contagious evolving pathogen that spreads rapidly among poultry sectors causes a threat to the poultry industry globally. H5N1 viruses of clade 2.3.4.4b have been widely circulating in wild-domestic birds and detected in Europe, Africa, North America, and Asia since October 2020. In this study, 40 tissue and swabs specimens were collected from 25 poultry flocks of different localities in New Valley Governorate, Egypt during 2023-2025. The collected samples were tested for H5N1, H9N2, H5N8, and H6N2 AIV subtypes as well as other pathogens such as NDV, IBV, ILT, and IBDV. To the best of our knowledge, this is first detection of HPAI-H5N1 of clade 2.3.4.4b viruses in broilers in New Valley Governorate, Egypt. Five positive H5N1 specimens were isolated in specific-pathogen-free-embryonated chicken eggs (SPF-ECE) with positive HA titers of 7–8 HA units. Using real-time reverse transcriptase polymerase chain reaction (RT-qPCR) assay targeting the M gene, only 4 samples were considered positive. Collectively, four samples were identified as positive for AIV of the H5N1 subtype through a one-step PCR assay, whereas the other samples were negative for other tested pathogens. Partial sequencing and the phylogenetic analysis of HA gene segment confirmed that the NewValley-1-H5N1-2023, and NewValley 2-H5N1-2024 isolates belonged to the H5N1 subtype of clade 2.3.4.4b. Our HPAI-H5N1 strains shared genetic similarity with the HPAI-H5N1 strains that reported in Europe, Asia, and Africa during 2021–2022. Currently, our H5N1 strains shows a genetic similarity to HPAIV-H5N1of clade 2.3.4.4b Egyptian isolates with nucleotide and amino acid identities percentage (96%-99%) meanwhile, they shared a low genomic relatedness (72%-84%) with commonly available vaccine. Genetically, the current H5N1 strains had R72S in the receptor binding sites of the HA protein of Egyptian H5N1 and had amino acid mutations in the HA immunogenic epitopes (A83D, T140A). Continuous monitoring and early determination of HPAI-H5N1 viruses in New Valley Governorate, Egypt as well as potentially updating the vaccine seed in a timely manner is essential to ensure the effectiveness of the vaccine as a control strategy of HPAI-H5 virus in the poultry industry.
Keywords
• Avian Influenza Virus
• H5N1
• HPAI
• Genetic Diversity
• Sequencing
• Clade 2.3.4.4b
Citation
Mohamed MK, Shehata MA, Mohamed MH, Abdelhafez MS (2025) Surveillance and Genetic Characterization of HPAI H5N1 in Poultry: Evidence from New Valley Governorate, Egypt. J Vet Med Res 12(2): 1285
INTRODUCTION
Avian Influenza Viruses (AIVs), particularly highly pathogenic H5 subtypes, pose severe threats to global poultry production and public health due to their genetic variability and pandemic potential [1]. AIVs are classified into 16 HA and 9 NA subtypes, with wild birds serving as reservoirs for their global spread [2,3]. While most AIVs are low pathogenic (LP), H5 and H7 subtypes often evolve into highly pathogenic (HP) strains, causing high mortality and economic losses [5]. Since the 1990s, HPAI H5 viruses have diversified into multiple clades (0-9) through mutation and reassortment. Notably, clade 2.3.4.4b H5N1 emerged in 2020, replacing earlier clades and spreading across continents via wild birds [6-8]. This clade exhibits broad host tropism, including mammals, raising concerns about zoonotic transmission. Egypt, situated on major migratory bird flyways, has faced endemic HPAI H5N1 (clade 2.2.1) since 2006 [9-12]. By 2016–2018, H5N8 (clade 2.3.4.4b) replaced H5N1 as the dominant subtype, challenging existing vaccines [13-15]. Egypt also reports the highest human H5N1 cases globally, underscoring ongoing zoonotic risks [16,17]. Despite vaccination campaigns, persistent outbreaks suggest antigenic mismatch between vaccines and circulating strains [18,19]. Genetic surveillance in understudied regions like Egypt’s New Valley Governorate is critical for outbreak control. This study reports the first detection of HPAI H5N1 clade 2.3.4.4b in New Valley broilers (2023–2025), analyzing its genetic divergence from vaccine strains and evolutionary trends.
MATERIALS AND METHODS
Ethical Approval
This study was approved by the New Valley Research Ethics Committee, Faculty of Veterinary Medicine, New Valley University (Approval No. 04-2024-100313), in compliance with animal experimentation guidelines AAIVE ver.2 2022.
Sample Collection
Source: Between 2023 and 2025, 40 pooled samples (tracheal/cloacal swabs and tissues) were collected from 25 broiler flocks (20–35 days old) in New Valley Governorate, Egypt. Flocks exhibited high mortality/ morbidity with respiratory/neurological signs. Necropsy findings such as cyanotic combs/wattles, facial intestinal/bursal proventricular petechiae were documented.Vaccination Status: Some flocks received inactivated H5N1 vaccines (Table 1).
Table 1: Epidemiological Data of Examined Flocks
|
Parameter |
Details |
|
Governorate |
New Valley, Egypt |
|
Flock Type |
Broilers (20–35 days) |
|
Vaccination |
Inactivated H5N1 (mixed coverage) |
|
Samples Collected |
Tracheal/cloacal swabs, tissues (n=40) |
Sample Processing
Tissue samples from brain, pancreas, kidney, lung, and trachea were homogenized in PBS (pH 7.4) with 10% antibiotics, centrifuged (3,000 rpm, 15 min), and supernatants stored at -80°C for analysis.
Virus Isolation and HA/HI Assays
Embryonated Eggs: Supernatants (0.2 mL) inoculated into 9-11day old SPF eggs (Nile SPF, Egypt) via allantoic cavity, incubated at 37°C for 4 days. HA Test: Allantoic fluid tested with 1% chicken RBCs. HA-positive samples stored at -80°C. HI Test: Confirmed H5N1 using monospecific antisera (H5N1, H5N8, NDV).
Molecular Detection
RT-qPCR: HA-positive samples screened for H5/H6/H9 and N1/N2/N8 subtypes (Verso 1-step™ Kit, Thermo Fisher) using primers/probes (Table 2). Co-infection Screening: Tested for IBV, NDV, ILTV, and IBDV.
Table 2: Primer and probe sets used for virus identification, subtyping, and sequencing in collected samples
|
ID |
Forward Primer (F) |
Reverse Primer (R) |
Probe Sequence |
|
AIV-M-gene |
AGATGAGTCTTCTAACCGAGGTCG (sep1) |
TGCAAAAACATCTTCAAGTCTCTG (sep2) |
FAM-TCAGGCCCCCTCAAAGCCGA-TAMRA |
|
AIV-H5 subtype |
ACATATGACTACCCACARTATTCAG (H5LH1) |
AGACCAGCTAYCATGATTGC (H5RH1) |
FAM-TCWACAGTGGCGAGTTCCCTAGCA-TAMRA |
|
AIV-H6 subtype |
CTTGGTGTGTATCAAATYCTTGC (IAV-H6-1666F) |
CATTGARCCATTTGARCACATCCA (IAV- H6-1776R) |
FAM-TATAGTACGGTATCGAGCAGYCT-MGB |
|
AIV-H9 subtype |
GGAAGAATTAATTATTATTGGTCGGTAC (For) |
GCCACCTTTTTCAGTCTGACATT (Rev) |
FAM-AACCAGGCCAGACATTGCGAGTAAGATCC-TAMRA |
|
AIV-N1 subtype |
TAYAACTCAAGGTTTGAGTCTGTYGCTTG (N1 forward) |
ATGTTRTTCCTCCAACTCTTGATRGTGTC (N1 reverse) |
FAM-TCAGCRAGTGCYTGCCATGATGGCA-TAMRA |
|
AIV-N2 subtype |
TGGACAGGGAACAACACTAAA (FN2) |
ACAAGCCTCCCATCGTAAAT (CN2) |
TXRED-CAAATGAAATGGAACACCCAACTCAT-BHQ23 |
|
AIV-N8 subtype |
TCCATGYTTTGGGTTGARATGAT (N8-1296F) |
GCTCCATCRTGCCAYGACCA (N8-1423R) |
FAM-TCHAGYAGCTCCATTGTRATGTGTGGAGT-TAMRA |
|
IBV |
ATGCTCAACCTTGTCCCTAGCA (AIBV-fr) |
TCAAACTGCGGATCATCACGT (AIBV-as) |
FAM-TTGGAAGTAGAGTGACGCCCAAACTTCA-TAMRA |
|
NDV |
TCCGGAGGATACAAGGGTCT (F+4839) |
AGCTGTTGCAACCCCAAG (F-4939) |
FAM-AAGCGTTTCTGTCTCCTTCCTCCA-TAMRA |
|
ILTV |
CCTTGCGTTTGAATTTTTCTGT (ILTVgCU771) |
TTCGTGGGTTAGAGGTCTGT (ILTVgCL873) |
FAM-CAGCTCGGTGACCCCATTCTA-MGBNFQ |
|
IBDV |
TCACCGTCCTCAGCTTACCCACATC (F/AUS GU) |
GGATTTGGGATCAGCTCGAAGTTGC (R/ AUS GL) |
Not provided |
Sequencing and Phylogenetics
congestion, and HA Gene Amplification: RT-PCR (Easyscript Kit) with primers KH1-Forward/KH3-Reverse (400 bp product). Sequencing: Purified PCR products (QIAquick Kit) sequenced (BigDye Terminator v3.1, ABI 3500xL). GenBank accessions: PV017774-PV017775. Phylogenetic Analysis: Aligned (Clustal W) and analyzed (MEGA 7.0, BioEdit) with reference strains (maximum likelihood, 1,000 bootstrap replicates).
RESULTS
Clinical and Pathological Findings
Prevalence: Avian influenza virus (AIV) was detected in 4 out of 25 examined broiler farms (16%) in New Valley Governorate during the period from 2023 to 2025.
Clinical Signs: Affected flocks showed respiratory signs including nasal discharge, coughing, and dyspnea; neurological signs including paresis, tremors, and opisthotonos; and digestive signs including greenish diarrhea. Morbidity reached 80%, and mortality ranged from 25% to 50%.
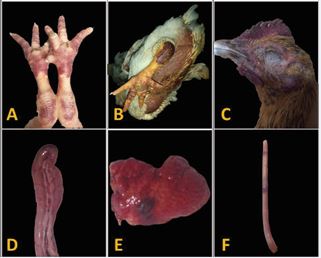
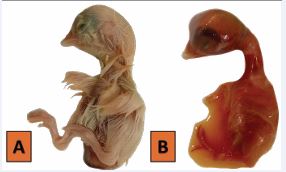
Gross Lesions: Postmortem findings included facial edema, pulmonary/intestinal congestion, proventricular hemorrhages, pancreatic necrosis, and subepicardial bleeding (Figure 1).
Figure 1: Gross lesions and clinical presentation of affected birds.
Virus Isolation and Serological Confirmation: Five H5N1 virus isolates were successfully recovered through inoculation into specific pathogen-free embryonated chicken eggs (SPF-ECE). The infected embryos exhibited characteristic pathological changes and died within 48 to 72 hours post-inoculation. Hemagglutination (HA) titers of the harvested allantoic fluids ranged from 7 to 8, indicating a high viral load (Figure 2).
To confirm antigenic specificity, the isolates were subjected to the hemagglutination inhibition (HI) assay using homologous H5N1 antiserum. The allantoic fluids demonstrated HI titers ranging from 5 to 7 log?, confirming the presence of H5-specific antibodies. No cross-reactivity was observed with H9 or Newcastle disease virus (NDV) antisera, indicating the absence of antigenic overlap and affirming the identity of the isolates as H5N1.
Molecular Characterization
Screening by real-time quantitative reverse transcription PCR (RT-qPCR) revealed that 4 out of 40 examined samples were positive for H5N1, as indicated by amplification of the matrix (M) gene. All positive samples were further tested for possible co-infections with Newcastle disease virus (NDV), infectious bronchitis virus (IBV), infectious laryngotracheitis virus (ILTV), and infectious bursal disease virus (IBDV), and all results were negative, confirming the absence of concurrent infections.Subtyping of the positive samples using H5 specific conventional RT-PCR produced amplicons of approximately 400 base pairs, further verifying the presence of H5 subtype avian influenza virus.
Genetic and Phylogenetic Analysis
Two H5N1 isolates, designated NewValley-1 H5N1-2023 and NewValley-2-H5N1-2024, were subjected to sequencing of the hemagglutinin (HA) gene and subsequently submitted to GenBank under accession numbers PV017774 and PV017775, respectively. Analysis of the HA cleavage site revealed the presence of the multibasic motif PLREKRRKR/GLF (amino acid positions 321–332), which is characteristic of highly pathogenic avian influenza viruses.Several notable mutations were identified, including R72S within the receptor binding site and A83D and T140A within antigenic site A, suggesting potential implications for host adaptation and immune escape. Phylogenetic analysis showed that both isolates belonged to clade 2.3.4.4b and exhibited high nucleotide identity (96–99%) with recent Egyptian strains from 2021 to 2022, such as A/ ibis/Egypt/RLQP-229S/2022. The isolates also clustered closely with contemporary H5N1 strains reported in Bangladesh, China, Lesotho, and the Czech Republic. In contrast, sequence homology with older Egyptian isolates (2010–2017) and currently used vaccinal strains was relatively low, ranging between 72% and 84%, indicating significant genetic divergence.
DISCUSSION
Highly Pathogenic Avian Influenza (HPAI) H5N1 virus was initially identified in 1996 in the Chinese poultry sector and subsequently disseminated globally through migratory birds, leading to devastating outbreaks in poultry and sporadic human infections [2-18]. Due to extensive circulation and accumulation of non-synonymous adaptive mutations in the viral surface proteins (antigenic drift), H5-type avian influenza viruses (AIVs) have evolved into nine major clades with numerous subclades. Of particular concern are the H5Nx viruses of clade 2.3.4.4, which include several emerging subclades (a–h) and have been increasingly reported [20].Since 2020, variants belonging to clade 2.3.4.4b have become the most predominant HPAI H5N1 strains, causing significant morbidity and mortality in wild and domestic bird populations, and posing considerable zoonotic risks [20-22]. In Egypt, multiple outbreaks of viral diseases, including AIV, have been documented [49–51]. Despite the implementation of routine vaccination programs, Egypt has remained an endemic region for AIVs since the virus was first reported in February 2006, with clade 2.2 HPAI H5N1 becoming endemic by 2008 [23,24].In late 2021, clade 2.3.4.4b H5N1 strains were introduced into Egypt via migratory birds and are currently co-circulating with H5N8 strains of the same clade, contributing to the ongoing viral evolution and environmental contamination [9-16]. The present study aimed to isolate and genetically characterize HPAI H5N1 viruses of clade 2.3.4.4b in New Valley Governorate, Egypt, during the period 2023–2025.Our findings demonstrated a 16% prevalence rate of AIV among sampled flocks, which exhibited marked clinical signs and post-mortem lesions, with associated mortality rates ranging from 25% to 50%. These clinical outcomes are consistent with previous reports [17-25]. The persistence of H5N1 in vaccinated flocks may be attributed to various factors, including improper vaccination practices, suboptimal vaccine coverage, limitations related to the vaccine seed strain and antigen content, as well as host-related variables such as species, age, immune status, and co-infections [6-17].Following inoculation of specific-pathogen-free embryonated chicken eggs (SPF-ECEs), embryos exhibited acute hemorrhagic and congestive lesions alongside positive HA titers, consistent with earlier findings [26,27]. RT-qPCR was employed as a rapid and sensitive tool for molecular detection, providing valuable epidemiological insights and phylogenetic data [28,29]. Among tested samples, only four were confirmed positive for HPAI H5N1, with no co-infections detected with NDV, IBV, ILT, or IBDV, in line with previous studies [7-30].Genetic characterization revealed that the two isolates (NewValley-1-H5N1-2023 and NewValley-2-H5N1-2024) belong to HPAI H5N1 clade 2.3.4.4b and exhibit high nucleotide and amino acid sequence identity (97%) with strains circulating in Africa, Asia, and Europe during 2021–2022. Phylogenetic analyses indicated strong genetic relatedness with strains from China, Lesotho, the Czech Republic, and Benin, suggesting a common introduction route via migratory birds along the Black Sea/Mediterranean flyway [31-34].Mutational analysis of the HA gene identified key amino acid substitutions (R72S, A83D, T140A) within immunogenic epitopes, underscoring the necessity for continuous genetic surveillance and periodic updates of vaccine strains to maintain efficacy. The HA cleavage site motif (PLREKRRKRGLF) aligns with previously reported Egyptian strains, confirming the highly pathogenic nature of the isolates [35,36].Historical data from 2014 to 2017 indicate the prior circulation of HPAI H5N1 clade 2.2.1.2 in New Valley and other Upper Egypt governorates [37], while the current isolates align more closely with clade 2.3.4.4b strains from 2021–2022, with 96%–99% identity. Additionally, a 95% 97% sequence similarity was observed with H5N8 strains (clade 2.3.4.4b) previously reported in Egypt between 2018 and 2021.Notably, sequence comparisons between the isolates and commonly used vaccine strains in Egypt revealed lower identity percentages (72%–84%), which may explain the suboptimal vaccine efficacy against emerging field strains. This observation aligns with prior studies reporting similar disparities (75.8%–90.7%) [9-16]. Overall, the divergence between circulating field strains and vaccine strains emphasizes the critical need for reformulation of vaccines to match the currently prevailing AIV genotypes.
CONCLUSIONS
This study provides the first report of the circulation of HPAI H5N1 viruses of clade 2.3.4.4b in New Valley Governorate, Egypt, during the period 2023–2025. The identified strains (NewValley-1-H5N1-2023 and NewValley-2-H5N1-2024) exhibited high genetic and phylogenetic similarity to contemporary H5N1 viruses reported in Europe, Asia, and Africa, indicating the probable role of migratory birds in their introduction and dissemination. These findings highlight the potential risk of further transmission to poultry populations across Egypt. Therefore, continuous surveillance, particularly at the interface between domestic and wild birds, is essential for early detection and prompt intervention before the virus becomes widely established or poses a zoonotic threat. The identified strains shared a high sequence identity (96%–99%) with recently circulating Egyptian H5N1 viruses of clade 2.3.4.4b but demonstrated limited homology (72%–84%) with commonly used vaccine strains. This genetic mismatch may contribute to vaccine failure and underscores the urgent need to re-evaluate current vaccination strategies, including the periodic updating of seed strains to improve vaccine efficacy.
ACKNOWLEDGEMENT
The authors thank the SciHub Office for providing professional English editing services for the manuscript.
REFERENCES
- Tripathi AK, Sendor AB, Sapra A. Avian Influenza. StatPearls. 2025.
- Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007; 25: 5637-5644.
- Capua I, Alexander DJ. Avian influenza infection in birds: a challenge and opportunity for the poultry veterinarian. Poult Sci. 2009; 88: 842-846.
- Xie XT, Yitbarek A, Astill J, Singh S, Khan SU, Sharif S, et al. Within-host model of respiratory virus shedding and antibody response to H9N2 avian influenza virus vaccination and infection in chickens. Infect Dis Model. 2021; 6: 490-502.
- Xie XT, Yitbarek A, Uddin Khan S, Sharif S, Poljak Z, Greer AL. A within-host mathematical model of H9N2 avian influenza infection and type-I interferon response pathways in chickens. J Theor Biol. 2020; 499: 110320.
- Arafat N, Abd El Rahman S, Naguib D, El-Shafei RA, Abdo W, Eladl AH. Co-infection of Salmonella enteritidis with H9N2 avian influenza virus in chickens. Avian Pathol. 2020; 49: 496-506.
- Arafat N, Eladl AH, Marghani BH, Saif MA, El-Shafei RA. Enhanced infection of avian influenza virus H9N2 with infectious laryngeotracheitis vaccination in chickens. Vet Microbiol. 2018; 219: 8-16.
- Ranjbar VR, Mohammadi A, Dadras H. Infectious bursal disease virus suppresses H9N2 avian influenza viral shedding in broiler chickens. Br Poult Sci. 2019; 60: 493-498.
- Abdelwhab EM, Abdel-Moneim AS. Epidemiology, ecology and gene pool of influenza a virus in Egypt: will Egypt be the epicentre of the next influenza pandemic? Virulence. 2015; 6: 6-18.
- Hassan KE, Saad N, Abozeid HH, Shany S, El-Kady MF, Arafa A, et al. Genotyping and reassortment analysis of highly pathogenic avian influenza viruses H5N8 and H5N2 from Egypt reveals successive annual replacement of genotypes. Infect Genet Evol. 2020; 84: 104375.
- Kayali G, Kandeil A, El-Shesheny R, Kayed AS, Maatouq AM, Cai Z, et al. Avian Influenza A(H5N1) Virus in Egypt. Emerg Infect Dis. 2016; 22: 379-388.
- Naguib MM, Arafa AS, El-Kady MF, Selim AA, Gunalan V, Maurer- Stroh S, et al. Evolutionary trajectories and diagnostic challenges of potentially zoonotic avian influenza viruses H5N1 and H9N2 co- circulating in Egypt. Infect Genet Evol. 2015; 34: 278-291.
- Kapczynski DR, Swayne DE. Influenza vaccines for avian species. Curr Top Microbiol Immunol. 2009; 333: 133-152.
- Swayne DE. Principles for vaccine protection in chickens and domestic waterfowl against avian influenza: emphasis on Asian H5N1 high pathogenicity avian influenza. Ann N Y Acad Sci. 2006; 1081: 174-81.
- Swayne DE, Lee CW, Spackman E. Inactivated North American and European H5N2 avian influenza virus vaccines protect chickens fromAsian H5N1 high pathogenicity avian influenza virus. Avian Pathol. 2006; 35: 141-146.
- El-Shesheny R, Kandeil A, Mostafa A, Ali MA, Webby RJ. H5 Influenza Viruses in Egypt. Cold Spring Harb Perspect Med. 2021; 11: a038745.
- Kayali G, El-Shesheny R, Kutkat MA, Kandeil AM, Mostafa A, Ducatez MF, et al. Continuing threat of influenza (H5N1) virus circulation in Egypt. Emerg Infect Dis. 2011; 17: 2306-2308.
- Castro-Sanguinetti GR, González-Veliz R, Callupe-Leyva A, Apaza- Chiara AP, Jara J, Silva W, et al. Highly pathogenic avian influenza virus H5N1 clade 2.3.4.4b from Peru forms a monophyletic group with Chilean isolates in South America. Sci Rep. 2024; 14: 3635.
- Mosaad Z, Elhusseiny MH, Zanaty A, Fathy MM, Hagag NM, Mady WH, et al. Emergence of Highly Pathogenic Avian Influenza A Virus (H5N1) of Clade 2.3.4.4b in Egypt, 2021-2022. Pathogens. 2023; 12: 90.
- Tian J, Bai X, Li M, Zeng X, Xu J, Li P, et al. Highly Pathogenic Avian Influenza Virus (H5N1) Clade 2.3.4.4b Introduced by Wild Birds, China, 2021. Emerg Infect Dis. 2023; 29: 1367-1375.
- Ge Z, Gu M, Cai T, Liu K, Gao R, Liu D, et al. Phylogenetic tracing and biological characterization of a novel clade 2.3.2.1 reassortant of H5N6 subtype avian influenza virus in China. Transbound Emerg Dis. 2021; 68: 730-741.
- Zhang X, Yang Y, Han X, Wei D, Niu B, Huang Q, et al. Unique phenomenon of H5 highly pathogenic avian influenza virus in China: co-circulation of Clade 2.3.4.4b H5N1 and H5N6 results in diversity of H5 Virus. Emerg Microbes Infect. 2025; 14: 2502005.
- Ali M, Yaqub T, Shahid MF, Wong FY, Mukhtar N, Naeem M, et al. Genetic Characterization of Highly Pathogenic Avian Influenza A(H5N8) Virus in Pakistani Live Bird Markets Reveals Rapid Diversification of Clade 2.3.4.4b Viruses. Viruses. 2021; 13: 1633.
- El-Shesheny R, Moatasim Y, Mahmoud SH, Song Y, El Taweel A, Gomaa M, et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Clade 2.3.4.4b in Wild Birds and Live Bird Markets. Egypt. Pathogens. 2022; 12: 36.
- Yang XY, Gong QL, Li YJ, Ata EB, Hu MJ, Sun YY, et al. The global prevalence of highly pathogenic avian influenza A (H5N8) infection in birds: A systematic review and meta-analysis. Microb Pathog. 2023; 176: 106001.
- Gonzales JL, Pritz-Verschuren S, Bouwstra R, Wiegel J, Elbers ARW, Beerens N. Seasonal risk of low pathogenic avian influenza virus introductions into free-range layer farms in the Netherlands. Transbound Emerg Dis. 2021; 68: 127-136.
- Stephens CB, Spackman E, Pantin-Jackwood MJ. Effects of an H7 Highly Pathogenic and Related Low Pathogenic Avian Influenza Virus on Chicken Egg Production, Viability, and Virus Contamination of Egg Contents and Surfaces. Avian Dis. 2020; 64: 143-148.
- Spackman E. Avian Influenza Virus Detection and Quantitation by Real-Time RT-PCR. Methods Mol Biol. 2020; 2123: 137-148.
- Spackman E, Suarez DL. Type A influenza virus detection and quantitation by real-time RT-PCR. Methods Mol Biol. 2008; 436: 19- 26.
- Hill NJ, Smith LM, Muzaffar SB, Nagel JL, Prosser DJ, Sullivan DJ, et al. Crossroads of highly pathogenic H5N1: overlap between wild and domestic birds in the Black Sea-Mediterranean impacts global transmission. Virus Evolution. 2021; 7: veaa093.
- Nagy A, Machova J, Hornickova J, Tomci M, Nagl I, Horyna B, et al. Highly pathogenic avian influenza virus subtype H5N1 in Mute swans in the Czech Republic. Vet Microbiol. 2007; 120: 9-16.
- Parvin R, Nooruzzaman M, Kabiraj CK, Begum JA, Chowdhury EH,Islam MR, et al. Controlling Avian Influenza Virus in Bangladesh: Challenges and Recommendations. Viruses. 2020; 12: 751.
- Pulit-Penaloza JA, Brock N, Belser JA, Sun X, Pappas C, Kieran TJ, et al. Highly pathogenic avian influenza A (H5N1) virus of clade2.3.4.4b isolated from a human case in Chile causes fatal disease and transmits between co-housed ferrets. Emerg Microbes Infect. 2024; 13: 2332667.
- Sanogo IN, Djegui F, Akpo Y, Gnanvi C, Dupré G, Rubrum A, et al. Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus in Poultry, Benin, 2021. Emerg Infect Dis. 2022; 28: 2534-2537.
- Wessels U, Abdelwhab EM, Veits J, Hoffmann D, Mamerow S, Stech O,et al. A Dual Motif in the Hemagglutinin of H5N1 Goose/Guangdong- Like Highly Pathogenic Avian Influenza Virus Strains Is Conserved from Their Early Evolution and Increases both Membrane Fusion pH and Virulence. J Virol. 2018; 92: e00778-18.
- Yang J, Qureshi M, Kolli R, Peacock TP, Sadeyen JR, Carter T, et al. The haemagglutinin gene of bovine-origin H5N1 influenza viruses currently retains receptor-binding and pH-fusion characteristics of avian host phenotype. Emerg Microbes Infect. 2025; 14: 2451052.
- Mohamed MK, Shahata MAE, Hafez MSA-e1 Mohamed MH, Soliman MA. Epidemiological and serological identification of avian influenza in middle Egypt. New Valley Veterinary J. 2025; 6: 1-6.