The Community Structure and Microbial Linkage of Rumen Ciliate, Methanogens and Fungi from Tashkurgan Sheep
- 1. College of Life and Geographic Sciences, Kashi University, China
- 2. Xinjiang Key Laboratory of Biological Resources and Ecology of Pamirs Plateau, China
ABSTRACT
Methane is one of the greenhouse gases. Methanogens can produce methane by metabolites of rumen microorganisms such as fungi and ciliates, that is, and the emission of methane in the process of culture is closely related to rumen microbial community. In this study, high-throughput sequencing technology was used to investigate the methanogen, ciliate and fungi communities and associations among methanogen and ciliate or fungi based on the abundances of genera and species. The results showed that Methanobacteriales and Cyclotrichida are dominant order of methanogen and ciliate respectively, and Neocallimastigomycota and Ascomycota are dominant phylum of fungi. In this study, we observed significant positive correlation between methanogens and ciliates, as well as positive and negative correlation between fungi and ciliates observed. Our results suggest that community structure of methanogens and fungi of Tashkurgan sheep rumen is consistent with other studies and ciliate community structure is different from other studies. Associations among methanogen and ciliate or fungi is related to the fermentation principle of methanogen, but it need to previous study that the mechanism of the correlation of between methanogen with fungi and ciliate identified in this study.
KEYWORDS
- Ruminant
- Animal husbandry
- Pamirs Plateau
CITATION
Wang Y, Tao Z (2024) The Community Structure and Microbial Linkage of Rumen Ciliate, Methanogens and Fungi from Tashkurgan Sheep. J Vet Med Res 11(2): 1270.
INTRODUCTION
CO2, CH4, N2O and F-gases are classified as greenhouse gases (GHG) because they absorb infrared radiation and cause greenhouse effect [1,2]. According to the report, global GHG emissions in 2022 reached an all-time high, of which CO2 accounted for 71.6%, CH4 21.0%, N2O 4.8% and F-gases 2.6%, and compared with 1990 and 2005 agricultural GHG emissions increased by 21% and 15%. This points to the need for more effective strategies to reduce GHG emissions [2]. The greenhouse effect of CH4 is 20-30 times that of CO2 [3]. Rumen of ruminant has microbial groups such as archaea, bacteria, fungi and protozoa, and the methanogen, belong to archaea, canmetabolize CO2, H2, acetic acid, methanol, methylamine, methylsulfide produced by the fermentation of bacteria, protozoa and fungi to produce methane [4-7]. Therefore, there was a close relationship between methanogenesis mechanism and methanogen community structure. Community structure of rumen methanogens was affected by dietary diet and seasons [8,9], but animal diet is the main driver of the changes in the rumen microbiota [10].
Tashkurgan sheep are mainly distributed in Tashkurgan Tajik Autonomous County, Kashi, Xinjiang, China. It has the advantages of adapting to high altitude and cold natural environment, strong resistance, large body and good meat production performance [11]. It is one of the main cultured species in Tashkurgan Tajik Autonomous County, and feed is mainly composed of cottonseed. Tashkurgan sheep′s live particularity environment and feed particularity food, but few researchers have considered methane emissions from the production of Tashkurgan sheep. In order to provide basic dates for the research of rumen methanogenesis mechanism of Tashkurgan sheep and finding ways to reduce methane emission in breeding. Present study, the community structure of methanogen, ciliate and fungi of Tashkurgan sheep rumen was analyzed by high-throughput sequencing, as well as the relationship between methanogen and ciliate or fungi provided.
MATERAL AND METHODS
DNA was extracted mainly using rumen contents from ten Tashkurgan sheep (MY01-MY10) in summer, and all samples were stored in the -80 refrigerator. Total genomic DNA of specimens was extracted using E.Z.N.A™ Mag-Bind Soil DNA Kit and the standard protocol of the Kit was used. The primers used in this study include: amplification of the 16sRNA of methanogens using GU1ST-340F/GU1ST-1000R and 349F/806R, amplification of the 18sRNA of protozoa using 18SV4F/ 18SV4R, amplification of the ITS of fungi using ITS1F/ ITS2 (Table 1).
Table 1: PCR primers used for methanogens, protozoa and fungi.
|
Primers |
Sequence |
Notes |
|
GU1ST-340F |
CCCTAYGGGGYGCASCAG |
|
|
GU1ST-1000R |
GGCCATGCACYWCYTCTC |
|
|
349F |
GYGCASCAGKCGMGAAW |
The primers were synthesized by Sangon Biotech (China). |
|
806R |
GGACTACVSGGGTATCTAAT |
|
|
ITS1F |
CTTGGTCATTTAGAGGAAGTAA |
|
|
ITS2 |
GCTGCGTTCTTCATCGATGC |
|
|
18SV4F |
GGCAAGTCTGGTGCCAG |
|
|
18SV4R |
ACGGTATCTRATCRTCTTCG |
|
The PCR products were shipped to the Sangon Biotech (China) for further processing. Use software Cutadapt 1.18 to remove the 3 ‘end sequencing primer connector from the original data; based on the overlap relationship between PE reads, PEAR 0.9.8 software concatenates paired reads into a sequence; distinguish sample sequences based on barcode sequences and primer sequences of each sample; use software PRINSEQ 0.20.4 to control and filter sequence quality. Perform OUT clustering analysis using software Usearch 11.0.667, and compare RDP database with RDP classifier 2.12 for taxonomic analysis; Perform alpha diversity index analysis using software motif 1.43.0, perform rafefaction analysis using motif 1.43.0, and plot using R 3.6.0 software to detect sequencing depth; based on taxonomic information, conduct statistical analysis of community structure at various classification levels and use Origin 2019 to plot, correlation analysis with software IBM Spss statistics.
RESULTS
Rumen Methanogen, Ciliate and Fungi Communities
80055-117465 high quality sequences mapped to Archaea 16s RNA gene. Three orders, four families and sis genera or subgenera of methanogen is identified (Table 2).
Table 2: The list of ethanogen.
|
Assigned taxa |
Assigned taxa |
Assigned taxa |
|
Methanobacteriales |
Methanomassiliicoccales |
Methanosarciniales |
|
Methanobacteriaceae |
Methanomethylophilaceae |
Methanosaetaceae |
|
Methanosphaera |
Candidatus Methanogranum |
Methanosaeta |
|
Methanobrevibacter |
Candidatus Methanomethylophilus |
Methanosarcinaceae |
|
Methanobrevibacter olleyae |
|
Methanimicrococcus |
|
Methanobrevibacter wolinii |
|
|
Table 3: The list of order and families of ciliate.
|
Assigned taxa |
Assigned taxa |
Assigned taxa |
|
Choreotrichida |
Nassulida |
Sporadotrichida |
|
Strombidinopsidae |
Nassulidae |
Trachelostylidae |
|
Cyclotrichida |
Philasterida |
Tintinnida |
|
Mesodiniidae |
Uronematidae |
Codonellidae |
|
Euplotida |
Prorodontida |
Tintinnidae |
|
Aspidiscidae |
Balanionidae |
Codonellopsidae |
|
Urostylida |
Plagiocampidae |
|
|
Holostichidae |
Norank_Spirotrichea |
|
|
Pseudokeronopsidae |
Strombidiidae |
|
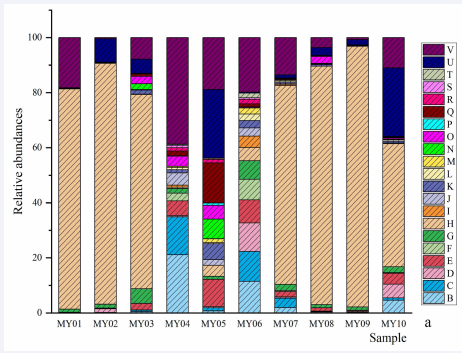
Methanobacteriales and Methanomassiliicoccales are dignosied in each sample, and Methanobacteriales is dominant order. Methanobacteriaceae and Methanomethylophilaceae can be identify in all sample and the family Methanobacteriaceae is dominant. Methanobrevibacter, Methanosphaera and Candidatus Methanomethylophilus can be identify in each sample and the genus Methanobrevibacter is dominant. The species Methanobrevibacter sp. is the dominant in all sample. The relative abundances of genus or subgenus and species are provided (Figure 1).
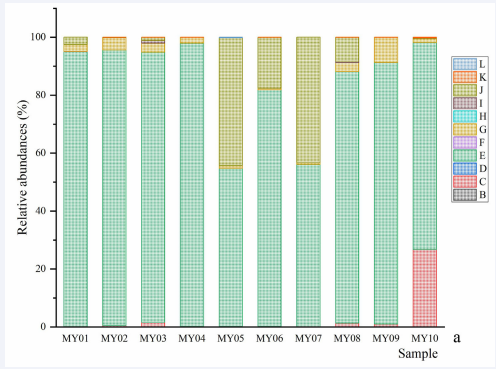
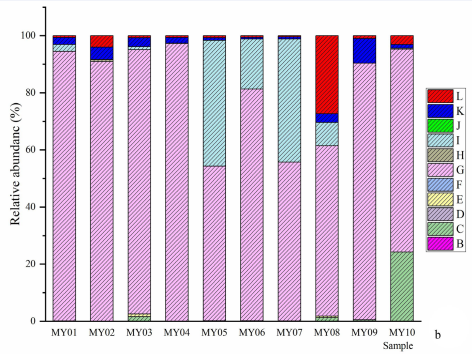
Figure 1a Genera and species of methanogen. a, relative abundances of genera and subgenera: B, Candidatus Methanogranum; C, Candidatus Methanomethylophilus; D, Methanimicrococcus; E, Methanobrevibacter; E, Methanosaeta; F, Methanosphaera; G, norank Bathyarchaeia; H, norank Methanobacteriaceae; I, norank Methanomethylophilaceae; J, unclassified Archaea; K, unclassified Methanomethylophilaceae. b, relative abundances of species: B, Bathyarchaeia sp.; C, Candidatus Methanomethylophilus sp.; D, Methanimicrococcus sp.; E, Methanobacteriaceae sp.; F, Methanobrevibacter olleyae; G, Methanobrevibacter sp.; H, Methanobrevibacter wolinii; I, Methanomethylophilaceae sp.; K, Methanosaeta sp.; L, Methanosphaera sp.; M, others. P=0.038), and between the species Methanosphaera sp. and Pecoramyces ruminantium (r=0.709, P=0.022); the negative correlations were observed between the species Methanosphaera sp. and some species of fungi, as well as between Caecomyces churrovis and Methanobrevibacter sp. (r=-0.661, P=0.038).
Figure 1b Genera and species of methanogen. a, relative abundances of genera and subgenera: B, Candidatus Methanogranum; C, Candidatus Methanomethylophilus; D, Methanimicrococcus; E, Methanobrevibacter; E, Methanosaeta; F, Methanosphaera; G, norank Bathyarchaeia; H, norank Methanobacteriaceae; I, norank Methanomethylophilaceae; J, unclassified Archaea; K, unclassified Methanomethylophilaceae. b, relative abundances of species: B, Bathyarchaeia sp.; C, Candidatus Methanomethylophilus sp.; D, Methanimicrococcus sp.; E, Methanobacteriaceae sp.; F, Methanobrevibacter olleyae; G, Methanobrevibacter sp.; H, Methanobrevibacter wolinii; I, Methanomethylophilaceae sp.; K, Methanosaeta sp.; L, Methanosphaera sp.; M, others.
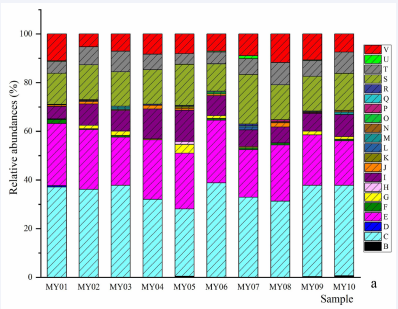
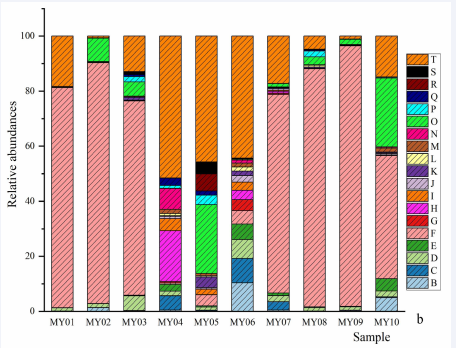
Figure 2a Genera and species of ciliates. a, relative abundances of genera: B, Apokeronopsis; C, Askenasia; D, Aspidisca; E, Balanion; E, Eutintinnus; F, Holosticha; G, Laackmanniella; H, Mesodinium; I, Nassula; J, Paraholosticha; K, Parallelostrombidium; L, Parastrombidinopsis; M, Plagiocampa; N, Pseudokeronopsis; O, Spirotrachelostyla; P, Stenosemella; R, Strombidinopsis; S, Strombidium; T, Tintinnopsis; U, Uronema, V, norank Ciliophora. b, relative abundances of species: B, Apokeronopsis crassa; C, Askenasia sp. LWW2010032604; D, Aspidisca fusca; E, Balanion planctonicum; F, Eutintinnus tubulosus; G, Holosticha bradburyae; H, Laackmanniella prolongata; I, Mesodinium rubrum; J, Mesodinium pulex; K, Nassula sp. LHA07091204; L, Paraholosticha muscicola; M, Parallelostrombidium paralatum; N, Parastrombidinopsis minima; O, Plagiocampa sp.; P, Pseudokeronopsis rubra; Q, Spirotrachelostyla tani; R, Stenosemella sp.; S, Strombidinopsis sp.; T, Strombidium biarmatum; U, Strombidium sp.1; V, Strombidium sp.2; W, Strombidium guangdongense; X, Tintinnopsis lobiancoi; Y, Tintinnopsis ventricosoides; Z, Tintinnopsis sp. JG-2011a; AA, Uronema nigricans; AB, norank Ciliophora.
Figure 2b Genera and species of ciliates. a, relative abundances of genera: B, Apokeronopsis; C, Askenasia; D, Aspidisca; E, Balanion; E, Eutintinnus; F, Holosticha; G, Laackmanniella; H, Mesodinium; I, Nassula; J, Paraholosticha; K, Parallelostrombidium; L, Parastrombidinopsis; M, Plagiocampa; N, Pseudokeronopsis; O, Spirotrachelostyla; P, Stenosemella; R, Strombidinopsis; S, Strombidium; T, Tintinnopsis; U, Uronema, V, norank Ciliophora. b, relative abundances of species: B, Apokeronopsis crassa; C, Askenasia sp. LWW2010032604; D, Aspidisca fusca; E, Balanion planctonicum; F, Eutintinnus tubulosus; G, Holosticha bradburyae; H, Laackmanniella prolongata; I, Mesodinium rubrum; J, Mesodinium pulex; K, Nassula sp. LHA07091204; L, Paraholosticha muscicola; M, Parallelostrombidium paralatum; N, Parastrombidinopsis minima; O, Plagiocampa sp.; P, Pseudokeronopsis rubra; Q, Spirotrachelostyla tani; R, Stenosemella sp.; S, Strombidinopsis sp.; T, Strombidium biarmatum; U, Strombidium sp.1; V, Strombidium sp.2; W, Strombidium guangdongense; X, Tintinnopsis lobiancoi; Y, Tintinnopsis ventricosoides; Z, Tintinnopsis sp. JG-2011a; AA, Uronema nigricans; AB, norank Ciliophora.
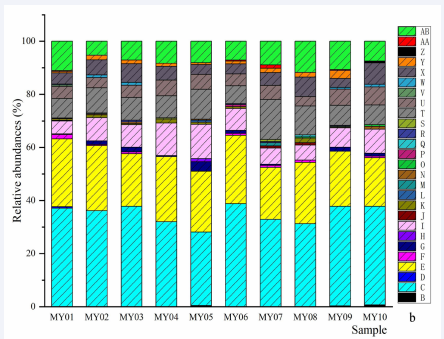
Figure 3a Genera and species of fungi. a, relative abundances of genera: B, Fusarium; C, Aspergillus; D, Blumeria; E, Alternaria; E, Paecilomyces; F, Kazachstania; G, Pecoramyces; H, Hyphopichia; I, Kodamaea; J, Pichia; K, Nigrospora; L, Davidiella; M, Caecomyces; N, Geotrichum; O, Lichtheimia; P, Penicillium; R, Vishniacozyma; S, Wallemia; T, Stemphylium; U, Neocallimastix; V, others. b, relative abundances of species: B, Blumeria graminis; C, Aspergillus ruber; D, Kazachstania humilis; E, Fusarium equiseti; F, Pecoramyces ruminantium; G, Hyphopichia burtonii; H, Fusarium oxysporum; I, Kodamaea ohmeri; J, Nigrospora sphaerica; K, Pichia kudriavzeviiI; L, Cladosporium aphidis; M, Trichothecium roseum; N, Aspergillus flavus; O, Neocallimastix frontalis; P, Geotrichum candidum; Q, Geotrichum klebahnii; R, Caecomyces churrovis; S, Candida ethanolica; T, Others.
Figure 3b Genera and species of fungi. a, relative abundances of genera: B, Fusarium; C, Aspergillus; D, Blumeria; E, Alternaria; E, Paecilomyces; F, Kazachstania; G, Pecoramyces; H, Hyphopichia; I, Kodamaea; J, Pichia; K, Nigrospora; L, Davidiella; M, Caecomyces; N, Geotrichum; O, Lichtheimia; P, Penicillium; R, Vishniacozyma; S, Wallemia; T, Stemphylium; U, Neocallimastix; V, others. b, relative abundances of species: B, Blumeria graminis; C, Aspergillus ruber; D, Kazachstania humilis; E, Fusarium equiseti; F, Pecoramyces ruminantium; G, Hyphopichia burtonii; H, Fusarium oxysporum; I, Kodamaea ohmeri; J, Nigrospora sphaerica; K, Pichia kudriavzeviiI; L, Cladosporium aphidis; M, Trichothecium roseum; N, Aspergillus flavus; O, Neocallimastix frontalis; P, Geotrichum candidum; Q, Geotrichum klebahnii; R, Caecomyces churrovis; S, Candida ethanolica; T, Others.
DISCUSSION
The survival of microorganisms in the rumen is based on the composition of chymus in the rumen, and the composition of chymus in different samples is different, so the relative abundances of microorganisms in different samples is different to some extent [12-15]. Methanogens include seven orders: Methanopyrales Methanococcales Methanobacteriales Methanomicrobiales Methanosarcinales, Methanocellales and Methanomassiliicoccales [16], and the number of Methanomassiliicoccales is usually second only to Methanobacteriales in the rumen [17,18], which is in agreement with present study. Genus Methanobrevibacter are dominant, which is also similar to results with previous study [19]. In present study, we found that there were great differences in the community structure of ciliates in different specie if Liu et al found that genus Polyplastron was the dominant genus in rumen of mountain [20], but it was not identified in present study or relative abundance was lower in Tan′s study [21]. The concentration and composition of ciliate in the rumen can be influenced by diet type, pH, supplements, concentrate and roughage level [22,23], but we could not find the literature on the correlation between ciliate community structure and ruminant species. The phylum Neocallimastigomycota and Ascomycota are dominant, which is in agreement with present study [24]. Methanobacteriales produces methane through reducing CO2; Methanomassiliicoccales produces methane through reducing methyl groups; Methanosarcinales produces methane through reducing CO2 methyl groups or acetic acid [25,26]. Protozoa play diverse roles in H2 production [27], which can be used as electron donors for methanogenic fermentation, so positive correlation observed between genus and species of ciliate protozoa with Candidatus Methanomethylophilus and Methanobrevibacter wolinii in this study. Fungi can produce H2, CO2, formic acid and acetic acid in fermentation, which provide electron donors or fermentation substrates for methanogens [28]. Therefore, the community structure of methanogens and fungi will establish a certain relationship. Nevertheless, the mechanism of the positive or cross-correlation between genera or species shown in this study needs further study.
CONCLUSION
The community structure of methanogens in Tashkurgan sheep feed with cottonseed is consistent with other studies, and the fungi at the phylum level is also consistent with other studies, but the ciliate community structure is different from other studies. Associations among methanogen and ciliate or fungi is related to the fermentation principle of methanogen. They need to be further studied that differences between ciliate community structure and it need to previous study that the mechanism of the correlation of between methanogen with fungi and ciliate identified in this study.
FUNDING
This work was financially supported by Kashi University Level Project ((2023)2853) and Program for Innovative Research Team in Kashi University.
REFERENCES
- Li BC, Dong LF, Cheng SR, Diao QY. Regulation of Methane Emissions from Ruminants with Different Crude-to-Ratio Diets. J. Domestic Anim. Ecol. 2019; 40:1-6.
- Crippa M, Unisystems SA, Guizzardi D, Pagani F, Unisystems SA, Banja M, et al. GHG emissions of all world countries. Publications Office of the European Union. 2023.
- Gill M, Smith P, Wilkinson JM. Mitigating climate change: the role of domestic livestock. Animal. 2010; 4: 323-333.
- Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM, et al. The Genome Sequence of the Rumen Methanogen Methanobrevibacter ruminantium Reveals New Possibilities for Controlling Ruminant Methane Emissions. 2010; 5: e8926.
- Gruninger RJ, Puniya AK, Callaghan TM, Edwards JE, Youssef N, Dagar SS, et al. Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol Ecol. 2014; 90: 1-17.
- Lan W, Yang CL. Ruminal methane production: Associated microorganisms and the potential of applying hydrogen-utilizing bacteria for mitigation. Sci Total Environ. 2019; 654: 270-1283.
- Beauchemin KA, Ungerfeld EM, Eckard RJ, Wang M. Review: Fifty years of research on rumen methanogenesis: lessons learned and future challenges for mitigation. Animal. 2020; 14: 1-16.
- Martinez-Fernandez G, Jiao JZ, Padmanabha J, Denman SE, McSweeney CS. Seasonal and Nutrient Supplement Responses in Rumen Microbiota Structure and Metabolites of Tropical Rangeland Cattle. Microorganisms. 2020; 8: 1550.
- Rabee AE, Kewan KZ, El Shaer HM, Lamara M, Sabra, EA. Effect of olive and date palm by-products on rumen methanogenic community in Barki sheep. Microbiol. 2022; 8: 26-41.
- Carberry CA, Waters SM, Kenny DA, Creevey CJ. Rumen methanogenic genotypes differ in abundance according to host residual feed intake phenotype and diet type. Appl Environ Microbiol. 2014; 80: 586-594.
- CHINA NATIONAL COMMISSION OF ANIMAL GENETIC RESOURCES. Animal Genetic Resources in China Sheep and Goat. Beijing: China Agricultural Press. 2011.
- Janssen PH, Kirs M. Structure of the archaeal community of the rumen. Appl Environ Microbiol. 2018; 74: 3619-3625.
- Carberry CA, Waters SM, Kenny DA, Creevey CJ. Rumen methanogenic genotypes differ in abundance according to host residual feed intake phenotype and diet type. Appl Environ Microbiol. 2014; 80: 586-594.
- Rabee AE, Forster R, Elekwachi C, Sabra E, Lamara M. Comparative analysis of the metabolically active microbial communities in the rumen of dromedary camels under different feeding systems using total rRNA sequencing. PeerJ. 2020; 8: e10184.
- Wang Z, Elekwachi CO, Jiao JZ, Wang M, Tang SX, Zhou CS, et al. Investigation and manipulation of metabolically active methanogen community composition during rumen development in black goats. Sci Rep. 2017; 7: 422.
- Garcia JL, Patel BKC, Ollivier B. Taxonomic, phylogenetic, and ecological diversity of methanogenic Archaea. Anaerobe. 2000; 6: 205-226.
- Seedorf H, Kittelmann S, Janssen PH. Few highly abundant operational taxonomic units dominate within rumen methanogenic archaeal species in New Zealand sheep and cattle. Appl Environ Microbiol. 2015; 81: 986-995.
- Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, et al. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. 2013; 8: e47879.
- Li ZP, Zhang ZG, Xu C, Zhao JB, Liu HL, Fan ZY, et al. Bacteria and methanogens differ along the gastrointestinal tract of Chinese roe deer (Capreolus pygargus). 2014; 9: e114513.
- Liu Q, Chen Y, Deng JL, Ren ZH, Yang YY, Cao S, et al. Ruminal Ciliate Community Structure of Chuanzhong Black Goats: An Analysis Using High-Throughput Sequencing Technology. Chinese J Anim Nutr. 2017; 29: 1574-1581.
- Tan C, Ramírez-Restrepo CA, Shah AM, Hu R, Bell M, Wang ZS, et al. The community structure and microbial linkage of rumen protozoa and methanogens in response to the addition of tea seed saponins in the diet of beef cattle. J Anim Sci Biotechnol. 2020; 11: 272-281.
- Hook SE, Steele MA, Northwood KS, Wright ADG, McBride BW. Impact of high-concentrate feeding and low ruminal pH on methanogens and protozoa in the rumen of dairy cows. Microb Ecol. 2011; 62: 94-105.
- Goel G, Makkar HPS, Becker K. Effects of Sesbania sesban and Carduus pycnocephalus leaves and fenugreek (Trigonella foenum-graecum L.) seeds and their extracts on partitioning of nutrients from roughage and concentrate-based feeds to methane. Anim Feed Sci Technol. 2008; 147: 72-89.
- Zhang T. Effects of Corn Silage levels on rumen fluid microbiota and its metabolome in Holstein Heifers. Beijing: China Agricultural University. 2017.
- Wang K, Nan XM, Xiong BH, Jiang LS. Research advances on rumen methanogenesis in ruminants. Chinese J Anim Nutr. 2020; 32: 5013- 5022.
- Zhou Y, Li Y, Jin W, Cheng YF, Zhu WY. Progress of Methanomassiliicoccales in the rumen. Acta Microbiol sin. 2020; 60: 1-12.
- Kholif AE. A Review of Effect of Saponins on Ruminal Fermentation, Health and Performance of Ruminants. Vet Sci. 2023; 10: 4500.
- Gruninger RJ, Puniya AK, Callaghan TM, Edwards JE, Youssef N, Dagar SS, et al. Anaerobic fungi (Phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol. Ecol. 2014; 90: 1-17.