The Community Structure and Prevalence, Antimicrobial Susceptibility Pattern, and Associated Risk Factors for Salmonella Species and Escherichia coli from Raw Meat at Butchery Houses in Mekelle, Tigray, Northern Ethiopia
- 1. Department of Medical Microbiology and Immunology, Mekelle University, Ethiopia
- 2. Department of Veterinary Basic and Diagnostic Sciences, Mekelle University, Ethiopia
ABSTRACT
Salmonella species and Escherichia coli (E.coli) are important foodborne pathogens affecting humans and animals. They are among the most important causes of infection that are associated with the consumption of contaminated food. This study aimed to determine the prevalence, antimicrobial susceptibility patterns, and associated risk factors for Salmonella species and E. coli in raw meat from butchery houses in Mekelle, Northern Ethiopia. Demographic data and risk factors were collected using a predesigned questionnaire. Meat samples were collected aseptically from the butchery houses and transported using an icebox to Mekelle University, College of Veterinary Sciences for the isolation and identification of Salmonella species and E. coli. Antimicrobial susceptibility patterns were determined using the Kirby disc diffusion method. Data obtained were cleaned and entered into Statistical Package for the Social Sciences version 22 and logistic regression models with odds ratio were calculated. P-value <0.05 was considered as statistically significant. A total of 153 out of 384 (39.8%) of the meat specimens were found to be contaminated. The contamination of Salmonella species and E.coli were 15.6% (n=60) and 20.8%) (n=80), respectively. Mixed contamination (Salmonella species and E.coli) was observed in 13 (3.4%) of the analyzed. Poor washing hands regularly (AOR=8.37; 95% CI: 2.75-25.50) and not using gloves during meat handling (AOR=11. 28; 95% CI: (4.69 27.10) were associated with an overall bacterial contamination. About 100% of the tested isolates were sensitive to Ciprofloxacin, gentamicin, Co trimoxazole (Trimethoprim-Sulphamethoxazole),sulfisoxazole/Sulphamethoxazole, ceftriaxone, and trimethoprim and ciprofloxacin, gentamicin and norfloxacine of E. coli and Salmonella species, respectively while the resistance of amoxyclav_ amoxicillin (clavulanic acid) and erythromycin were both isolated bacteria species. The overall multidrug resistance pattern for Salmonella and E. coli was 51.4% (n=19) and 31.8% (14), respectively. Of the 153 (153/384) contaminated raw meat, 60 (15.6%) and 80 (20.8%) were contaminated by Salmonella species and E. coli, respectively. Poor handwashing practices and not using gloves during meat handling showed significant association with bacterial contamination. Multidrug-resistant showed in Salmonella species and E. coli were 19 (51.4%) and 14 (31.8%), respectively.
KEYWORDS
- Antimicrobial Susceptibility test
- Butchery houses
- Escherichia coli
- Raw meat
- Salmonella species
CITATION
Tadesse HA, Hagos DG, Kahsay AG, Abdulkader MA (2024) Prevalence, Antimicrobial Susceptibility Pattern, and Associated Risk Factors for Salmonella Species and Escherichia coli from Raw Meat at Butchery Houses in Mekelle, Tigray, Northern Ethiopia. J Vet Med Res 11(3): 1271.
ABBREVIATIONS
MDR: Multidrug-resistant; CLSI: Clinical laboratories Standards institute; SPSS: Statistical Package for Social Sciences.
INTRODUCTION
Food safety remains a concern of global human health [1]. The difficulties in securing optimal hygienic food handling practices in developing countries lead to food contamination [2]. Above two-thirds, (70%) of diarrheal diseases in developing countries are reported to be the consumption of contaminated food [1]. Food associated with animal origin is the cause of over 60% of human pathogens [2]. Meat can be infected or carry a wide range of microorganisms, which are potentially pathogenic for humans [3]. Most of the bacteria that contaminate the meat are zoonotic bacteria including E. coli and Salmonella species [4,5].
The annual estimated incidence of Salmonella species in the USA is more than 1.2 million illnesses, 23,000 hospitalizations, and 450 deaths [6] with the highest cost burden [7]. In Europe, infections caused by Salmonella species are the second leading cause of bacterial foodborne illness [8]. The estimated economic burden of human salmonellosis and E. coli strain could be as high as 3 billion euros per year as reported by the European Food Safety Authority [9].
As indicated in different studies in Ethiopia, the prevalence rate of Salmonella species and E.coli in raw meat samples was from 2.5% to 14.7% [10-16].
Indiscriminate use of antibiotics in livestock production, as well as human diseases in developing countries, lead to an increase in antimicrobial-resistant (AMR) bacteria [17]. The annual estimated death of individuals due to antimicrobial resistance in the USA and Europe is 23,000 [18] and 25,000 [19], respectively. The global death of individuals due to antimicrobial- resistant bacteria is estimated to be 700,000 [20].
Meat is highly vulnerable to microbiological hazards and therefore needs careful handling, transporting, and storing. Unhygienic conditions during the handling of raw meat during the meat value chain implied a possible risk of infection (Ministry of Agriculture (MOA), 2010) [21]. Salmonella species and E.coli are among the common bacteria that can contaminate the meat along the meat chain.
Therefore, due to the lack of data in the study area, this study was intended to carry out the prevalence, antimicrobial susceptibility pattern, and associated factors for Salmonella species and E. coli from raw meat at butchery houses in Mekelle, Tigray, Ethiopia.
MATERIALS AND METHODS
Study area and study design. The study was conducted in Mekelle, Tigray Regional State, Northern Ethiopia. Mekelle is the capital city of the regional state found located at 390291E and 130 301N latitudes and longitudes at a distance of 783 Km north of Addis Ababa. The capital city covers an area of 109 square kilometres with an elevation is 2,084 m/s above sea level. The climatic condition of the area is characterized by semi-arid weather with bimodal rainfall patterns, with an average annual rainfall of 479 to 650 mm. The annual average temperature is 20.9°C with an annual mean humidity of 75.4% [22]. A cross- sectional study was conducted from January to December 2019. All the butchery shops in Mekelle City were included in the study.
SAMPLING TECHNIQUE AND SIZE DETERMINATION
Sampling Technique
A consecutive sampling technique was employed to recruit the butchery houses. Raw meats from each of the butchery houses were collected by using a simple random sampling technique using lottery methods.
Sample Size Determination
The sample size was determined using a single proportion. The calculation was based on a prevalence of 50%, 5% desired absolute precision (or error), and a 95 % confidence interval using the formula.
n=Z2 p (1-p) calculate the correct number n= (1.96)2 * 50(1-0.05) = 384 / d2 (0.05)2
Where n=required sample size; p = expected prevalence and a desired absolute precision (d) of 0.05, Z-value=1.96. Therefore, a total of 384 samples of butchery houses used for this study were 384.
Data Collection and Sample Processing. Socio-demographic, hygiene, and sanitation practice data were collected from the individuals working in the butchery shops. From the study participants in the butchery houses.
Sample collection, handling, and transportation. Raw meat samples were collected in a labeled sterile bottle following aseptic techniques and were transported in a buffer peptone water broth in the icebox to Mekelle University, College of Veterinary Sciences, Microbiology and Public Health Laboratory for isolates of microbiological and antimicrobial susceptibility testing.
BACTERIAL ISOLATION
Salmonella Species
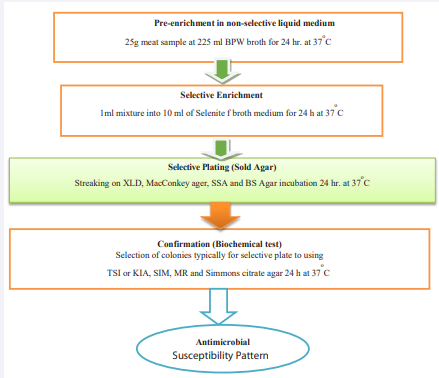
Salmonella species were isolated and identified according to the technique recommended by the International Organization for Standardization. Detection of salmonella species was performed using the standard guidelines from ISO 6579: 2002. This isolation and identification procedure involves principal stages including pre-enrichment, selective enrichment, selective plating, and conformation using biochemical tests [23].
Pre-Enrichment
Raw meat specimens were pre-enriched inappropriate amount of buffered peptone water and lactose broth in a (1:9) ratio or twenty-five grams of raw meat was placed in 225 ml of peptone water or lactose broth to produce high resuscitation rates for bacteria and promote intense growth. The sample mixture was shaken approximately for 2 minutes and was incubated at 37±1oC for 24 hours [24].
Selective Enrichment
Selenite F broth was used for selective enrichment purposes. About 1 ml of the pre-enriched broth was transferred into a tube containing 10 ml of Selenite F broth and was incubated at 37°C for 24 hours [24].
Isolation of Salmonella Species
Xylose lysine deoxycholate (XLD) agar, McConkey agar, salmonella and shigella agar (SSA), and bismuth sulfite (BS) agar plates were used for plating out and identification. A loop full of inoculums from Selenite broth cultures was inoculated into XLD, BS, SSA, and MacConkey agar plates and was incubated at 37OC for 24 hours. After incubation, the plates were examined for the presence of typical and suspect colonies. Typical colonies of Salmonella grown on XLD agar have a black center and a light transparent zone of reddish color due to the color change of the media while H2S negative variants grown on XLD agar are pink with a darker pink center. On BS agar, Salmonella colonies are brown, grey, or black, sometimes with a metallic sheen. Typical colonies of Salmonella on SSA are with a black center (spot black at center), 1 mm to 2 mm in diameter, and cause the color of medium to change to typical colonies or suspected colonies were selected from the selective plating media, streaked onto the surface of pre-dried nutrient agar plates and incubated at 37oC for 24 hrs then indicated to [Figure 1] [24].
Figure 1 Schematic procedure for Salmonella species isolation.
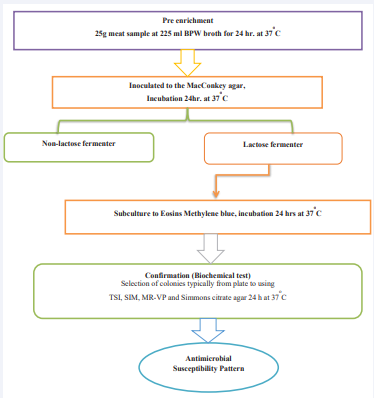
Escherichia Coli
Isolation of E.coli was conducted following standard procedure. Upon arrival to the laboratory, all pre-enriched buffered peptone water broth raw meat samples were subsequently inoculated to MacConkey agar and were incubated at 37 OC overnight bacterial growth was subjected to a lactose fermenter and non-lactose fermenter, and the lactose fermenter colony was sub-cultured to Eosin methylene blue (EMB) agar and were incubated at 37 °C for 24 hours. Colonies showing typical dark red to purple red with metallic sheen were taken as E.coli isolates then indicated in [Figure 2] [25].
Figure 2 Schematic procedure for Escherichia coli isolation.
Biochemical Tests
Identification of Salmonella species and E. coli was done using different biochemical tests including catalase, triple sugar iron (TSI) agar, Methyl red (MR), urease, SIM agar (sulfide, indole, and motility), and citrate tests. Colonies that showed red slant with yellow butt and H2 S production, Indole negative, methyl red positive, citrate positive, and urease negative were confirmed as Salmonella species whereas colonies that showed yellow slant and acid butt with no hydrogen sulfide, Indole positive, motile, methyl red positive, citrate negative and urease negative were confirmed as E.coli [25].
ANTIMICROBIAL SUSCEPTIBILITY TESTING
A modified Kirby-Bauer disc diffusion technique was used to perform an antimicrobial susceptibility test. The pure colony of E. coli and Salmonella species were tested in separate Mueller Hinton agar. With a sterile wire loop, five colonies of similar morphological type were transferred to a tube containing 3-5ml normal slain solution (NSS). E. coli and Salmonella species were tested for susceptibility to the following 16 antibiotics (OXOID, England): amoxicillin/clavulanic acid (30 μg), ceftriaxone (30 μg), chloramphenicol (30 μg), ciprofloxacin (30 μg), gentamycin (10 μg), kanamycin (30 μg), nalidixic acid (30 μg), nitrofurantoin (30 μg), streptomycin (10 μg), trimethoprim/sulfamethoxazole, and using the disk diffusion method according to guidelines set by the Clinical Laboratory Standards Institute [26]. E.coli and Salmonella species separate suspensions were prepared and matched with McFarland standard (0.5) and were seeded using an applicator cotton swab to the muller-hinton agar and put the paper-impregnated antibiotic disks within 15 minutes. Petri-dish was incubated at 37oC for 16-18 hrs. After 18 hrs incubation, each plate was examined, and the diameters of the complete inhibition zones were noted and measured using calipers and classified as sensitive, intermediate, and resistant by the Clinical Laboratory Standards Institute [26].
DATA QUALITY CONTROL
Data completeness, expired date of media and disks, and sterility test of media were performed before data collection and sample inoculation. Quality control (E. coli ATCC ATCC 25922) was used to check the performance of the media and antibiotic disks.
DATA MANAGEMENT AND ANALYSIS
Statistical Package for Social Sciences (SPSS) version 22 software for Windows was used. Descriptive analysis was presented using tables. Binomial and multinomial regression was used to determine association contamination rate and bacterial isolates were determined by calculating odds ratio and 95% confidence interval and P-value<0.05 was considered as statistically significant.
ETHICAL CLEARANCE
Ethical clearance was obtained from the ethical review board (ERC-1217/2019) of Mekelle University, College of Health Sciences. Study participants and/or their relatives were informed about the procedures and significance of the study. Consent was obtained from participants and owners of the butchery house. Each data results were kept confidential. All laboratory tests were free of any charge and results were communicated to relevant offices and community for beneficiary measures.
RESULTS
Socio-demographic characteristics of the total 384 study participants, 344 (89.6%) were males. The majority 329 (85.7%) of the study participants were in the age range of 31-40 years. The educational status of 109 (28.4%) and 108 (28.1%) of the study participants was college/university and high school, respectively whereas thirty-six of the participants (9.4%) were illiterates. The work experience of the majority of the participants, 240 (62.5%) was 2-5 years as indicated in Table 1.
Table 1: Socio-demographic characteristics of study participants working in the butchery houses in Mekelle, Tigray, Ethiopia, 2019.
|
Factors (Variable) |
Value |
Frequency (%) |
|
Age |
18-30 |
36 (9.4) |
|
|
31-40 |
329 (85.7) |
|
|
>==41 |
19 (5.0) |
|
Sex |
Female |
40 (10.4) |
|
|
Male |
344 (89.6) |
|
Education States |
Illiterate |
36 (9.4) |
|
|
Read and Write |
43 (11.2) |
|
|
Primary |
88 (22.9) |
|
|
High school |
108 (28.1) |
|
|
College/University |
109 (28.4) |
|
Work Experience |
<=1 |
19 (5.0) |
|
|
2-5 |
240 (62.5) |
|
|
6-10 |
106 (27.6) |
|
|
>=10 |
19 (5.0) |
Prevalence of Salmonella Species and E.coli from Raw meat Samples
A total of 384 butchery houses were included in the study. One raw meat sample was from each of the butchery houses. One hundred fifty-three (39.8%) of meat contaminants (Salmonella and E.coli) were recovered from the collected raw meat specimens. Sixty (15.6%) Salmonella species, eighty (20.8%) E.coli, and thirteen mixed contamination (3.4%) (Salmonella and E.coli) were revealed from the raw meat samples.
Associated Risk factors for meat contamination
Risk factors that showed significant association in multivariate logistic regression analysis of meat contamination were poor regularly washing hands (AOR=8.37; 95% CI: 2.75-25.50) and not using gloves during meat handling and selling (AOR=11. 28; 95% CI: (4.69-27.10) as indicated in Table 2.
Table 2: Risk factors associated with bacterial isolates from the contaminated meat samples in Mekelle, Tigray, Ethiopia, 2019.
|
Factors (Variable) |
Frequency (%) |
Bacterial contaminants ( Salmonella and E. coli ) |
||
|
COR (95%CI) P-Value |
AOR (95%CI) P-Value |
|||
|
Wash hands regularly |
|
|
|
|
|
|
Yes |
185 (48.2) |
Reference |
Reference |
|
No |
199 (51.8) |
84.5 (35.3- 202.6) 0.001 |
8.37 (2.75-25.50) 0.001 |
|
|
Using gloves |
|
|
|
|
|
|
Yes |
181( 47.1) |
Reference |
Reference |
|
|
No |
203 (52.9) |
43.3 (21.8-86.1) 0.001 |
11.28 (4.69—27.10) 0.001 |
|
Strict Separation b/n clean & dirty |
|
|
|
|
|
|
Yes |
189 (49.2) |
Reference |
Reference |
|
|
No |
195 (50.8) |
0.53 (0.35- 0.81) 0.003 |
0.38 (0.17–0.85) 0.019 |
|
Knife can transfer disease |
|
|
|
|
|
|
Yes |
185 (48.2) |
Reference |
Reference |
|
|
No |
199 (51.8) |
44.7 (21.97-90.85) 0.001 |
3.33 (1.16-9.56) 0.025 |
|
Cleaning equipment After work |
|
|
|
|
|
|
Yes |
184 (47.9) |
Reference |
Reference |
|
|
No |
200 (52.1) |
60.9 (28.02-132.5) 0.001 |
6.08 (2.08-17.82) 0.001 |
CI-Statistically significant at 95% confidence interval, p-value= 0.05%, COR-Crude Odds Ratio, AOR-Adjusted Odds Ratio.
Associated Risk Factors for Salmonella Species and Escherichia Coli Contamination
Among the associated risk factories of handling money with bare hand during raw meat selling (AOR=6.98; 95% CI: 2.46- 19.86), Cutting board can transfer (AOR=10.50; 95% CI: 2.49- 44.27) and not using gloves during meat handling (AOR=4.87; 95% CI: 1.13-21.06) were found to be significantly associated with Salmonella species and handling money with bare hand during raw meat selling (AOR=10.89; 95% CI: 3.48-34.05) and sources of water (AOR=9.67; 95% CI:4.22-22.16) and poor hand washing regularly (AOR=56.69; 95% CI: 11.95-268.88) were found to be significantly associated with E.coli as indicated in [Table 3].
Table 3: Risk factors associated with Salmonella species and E. coli isolates from contaminated raw meat in butchery houses of Mekelle, Tigray, Ethiopia.
|
Factors (Variable) |
Frequency (%) |
Salmonella species |
Escherichia coli |
||||
|
COR (95%CI) |
AOR (95%CI) |
COR (95%CI) |
AOR (95%CI) |
||||
|
Wash hands regularly |
|
|
|
|
|
||
|
|
Yes |
185 (48.2) |
Reference |
|
Reference |
|
|
|
No |
199 (51.8) |
24.57 (8.720-69.244)* |
1.49 (0.23-9. 59) |
83.73 (20.19-347.29)* |
56.69 (11.95-264.88)* |
||
|
Washing hand properly |
|
|
|
|
|
||
|
|
Yes |
184 (47.9) |
Reference |
|
Reference |
|
|
|
|
No |
200 (52.1) |
13.01 (5.76-29.41)* |
0.61 (0.12 – 3.04) |
21.21 (9.45-47.60)* |
1.19 (0.17-8.32) |
|
|
Using gloves |
|
|
|
|
|
||
|
|
Yes |
181 (47.1) |
Reference |
Reference |
Reference |
|
|
|
|
No |
203 (52.9) |
35.59 (10.93-115.87)* |
4.87 (1.13- 21.06)* |
18.88 (8.78-40.60)* |
1.05 (0.17-6.43) |
|
|
Using hot water to clean |
|
|
|
|
|
||
|
|
Yes |
189 (49.2) |
Reference |
|
Reference |
|
|
|
|
No |
195 (50.8) |
18.11 (7.08-46.31)* |
2.07 (0.46-9.33) |
29.12 (11.45-74.06)* |
0.61 (0.09-3.98) |
|
|
Cutting Board can transfer |
|
|
|
|
|
||
|
|
Yes |
181 (47.1) |
Reference |
|
Reference |
|
|
|
|
No |
203 (52.9) |
35.59 (10.93-115.87)* |
10.50 (2.49- 44.27)* |
42.14 (15.01-118.27)* |
1.65 (0.07-38.68) |
|
|
Cleaning equipment After work |
|
|
|
|
|
||
|
|
Yes |
184 (47.9) |
Reference |
|
Reference |
|
|
|
|
No |
200 (52.1) |
24.90 (8.84-70.18)* |
0.73 (0.12-4.34) |
40.27 (14.35-112.98)* |
2.42 (0.24-24.13) |
|
|
Sources of water |
|
|
|
|
|
||
|
|
Tap |
119 (30.99) |
Reference |
|
Reference |
|
|
|
Well |
265 (69.01) |
2.91 (1.69- 5.01)* |
0.47 (0.23-1.04) |
26.85 (14.18- 50.81)* |
9.67 (4.22- 22.16)* |
||
|
Equipment’s rested on dirty surfaces during working |
|
|
|
|
|
||
|
|
Yes |
174 (45.31) |
Reference |
|
Reference |
Reference |
|
|
|
No |
210 (54.7) |
3.09 (1.69-5.65)* |
1.04 (0.41-2.59) |
10.83 (5.24-22.41)* |
4.68 (1.69 - 12.93)* |
|
|
Handling money |
|
|
|
|
|
||
|
|
Cashier |
188 (48.96) |
Reference |
Reference |
Reference |
Reference |
|
|
Butcher with Bare hands |
196 (51.04) |
14.01 (5.49-35.80)* |
6.98(2.46- 19.86)* |
17.82 (7.54-42.14)* |
10.89 (3.48 - 34.05)* |
||
|
Cutting table |
|
|
|
|
|
||
|
|
Separate for difference |
188 (49) |
Reference |
Reference |
Reference |
Reference |
|
|
|
Single |
209 (54.4) |
0.20 (0.11-0.38)* |
0.16 (0.07- 0.33)* |
1.66 (1.02-2.68)* |
5.15 (2.11-12.57)* |
|
*Statistically significant at 95% confidence interval, COR-Crude Odds Ratio, AOR-Adjusted Odds Ratio.
Antimicrobial Susceptibility Test
Sixteen antimicrobial discs were used to assess the susceptibility pattern of isolates. All of the tested (n=44) E. coli isolates were sensitive to co-trimoxazole, sulfisoxazole/ sulphamethoxazole, trimethoprim, ciprofloxacin, gentamicin and ceftriaxone whereas their sensitivity to nalidixic acid, chloramphenicol, norfloxacin and nitrofurantoin were 93.2%) (n=41), 95.5% (n=42), 95.5% (n=42) and 84.1%) (n=37), respectively. The sensitivity of E. coli to doxycycline hydrochloride, cefotaxime, kanamycin, and streptomycin were 27 (61.4%), 25 (56.8%), 25 (56.8%), and 25 (56.8%), respectively and isolates were resistance to erythromycin, amoxyclav_amoxicillin, kanamycin, streptomycin, and cefotaxime were 33 (75%), 39(88.6%) and 19 (43.2%), respectively.
Similarly, all of the tested isolates (n=37) of Salmonella species were sensitive to gentamicin, norfloxacin and ciprofloxacin, whereas the sensitivity for nalidixic acid, sulfisoxazole/ Sulphametoxazole, trimethoprim, cefotaxime, doxycycline hydrochloride, ceftriaxone, and kanamycin were 35 (94.6%), 35 (94.6%), 35 (94.6%), 31 (83.8%), 31 (83.8%), 31 (83.8%), and 30 (81.8%), respectively and isolates were resistance to erythromycin, amoxyclav_amoxicillin and nitrofurantoin were 21 (56.8%), 32 (86.5%) and 16 (43.2%), respectively as indicated in Table 4.
Table 4: Antimicrobial susceptibility patterns of E. coli and Salmonella species isolates from contaminated raw meat in Butchery houses of Mekelle, Tigray, Ethiopia, 2019.
|
Antibiotic type |
Status of an antimicrobial agent against the isolates |
|||||
|
E. coli (n=44) |
Salmonella species (n=37) |
|||||
|
R (%) |
I (%) |
S (%) |
R (%) |
I (%) |
S (%) |
|
|
Nalidixic Acid |
- |
3 (6.8) |
41 (93.2) |
2 (5.4) |
- |
35 (94.6) |
|
Norfloxacine |
2 (4.5) |
- |
42 (95.5) |
- |
- |
37 (100) |
|
Chloramphenicol |
2 (4.5) |
- |
42 (95.5) |
3 (8.1) |
11 (29.7) |
23 (62.2) |
|
Trimethoprim |
- |
- |
44 (100) |
2 (5.4) |
- |
35 (94.6) |
|
Ciprofloxacin |
- |
- |
44 (100) |
- |
- |
37 (100) |
|
Erythromycin |
33 (75) |
5 (11.4) |
6 (13.6) |
21 (56.8) |
2 (5.4) |
14 (37.8) |
|
Gentamicin |
- |
- |
44 (100) |
- |
- |
37 (100) |
|
Kanamycin |
19 (43.2) |
- |
25 (56.8) |
- |
7 (18.2) |
30 (81.8) |
|
Streptomycin |
19 (43.2) |
- |
25 (56.8) |
11 (29.7) |
- |
26 (70.3) |
|
Sulphamethoxazole |
- |
- |
44 (100) |
- |
2 (5.4) |
35 (94.6) |
|
Ceftriaxone |
- |
- |
44 (100) |
4 (10.8) |
2 (5.4) |
31 (83.8) |
|
Co-trimoxazole |
- |
- |
44 (100) |
2 (5.4) |
- |
35 (94.6) |
|
Nitrofurantoin |
1 (2.3) |
6 (13.6) |
37 (84.1) |
16 (43.2) |
5(13.5) |
16 (43.3) |
|
Cefotaxime |
19 (43.2) |
- |
25 (56.8) |
5 (13.5) |
1 (2.7) |
31 (83.8) |
|
Doxycycline |
17 (38.6) |
- |
27 (61.4) |
6 (16.2) |
- |
31 (83.8) |
|
Amoxyclav_Amoxicillin |
39(88.6) |
3 (6.8) |
2 (4.6) |
32 (86.5) |
3 (8.1) |
2 (5.4) |
I= Intermediate, R= Resistance, S= Sensitive
Multidrug resistance of Salmonella and E.coli Isolates
Nineteen (51.4%) of the isolates of the Salmonella species and fourteen (31.8%) of the isolates of E. coli showed multidrug resistance (MDR) as indicated in [Table 5].
Table 5: Multiple drug resistance of Salmonella and E. coli isolates from contaminated raw meat in Butchery houses of Mekelle, Tigray, Ethiopia, 2019.
|
Anti-microbial |
Salmonella species (N=37) |
E. coli (N=44) |
||||||||||
|
Resistance pattern |
Isolates N (%) |
Resistance pattern |
Isolates N (%) |
|||||||||
|
Three |
E |
NIT |
DO |
|
|
1 (5.3) |
NX |
DO |
AMC |
|
|
2 (14.3) |
|
E |
NIT |
AMC |
|
|
5 (26.3) |
NX |
E |
AMC |
|
|
1 (7.1) |
|
|
C |
E |
AMC |
|
|
1 (5.3) |
CTX |
E |
AMC |
|
|
1 (7.1) |
|
|
S |
DO |
AMC |
|
|
1 (5.3) |
E |
DO |
AMC |
|
|
5 (35.7) |
|
|
TR |
E |
AMC |
|
|
1 (5.3) |
E |
S |
AMC |
|
|
1 (7.1) |
|
|
|
|
|
|
|
|
E |
CTX |
AMC |
|
|
1 (7.1) |
|
|
Four |
C |
E |
NIT |
AMC |
|
1 (5.3) |
C |
E |
DO |
AMC |
|
1 (7.1) |
|
E |
S |
DO |
AMC |
|
2 (10.5) |
E |
NIT |
DO |
AMC |
|
1 (7.1) |
|
|
C |
E |
NIT |
AMC |
|
1 (5.3) |
|
|
|
|
|
|
|
|
E |
S |
DO |
AMC |
|
1 (5.3) |
|
|
|
|
|
|
|
|
NA |
CTR |
CTX |
AMC |
|
1 (5.3) |
|
|
|
|
|
|
|
|
TR |
E |
NIT |
AMC |
|
1 (5.3) |
|
|
|
|
|
|
|
|
E |
NIT |
DO |
AMC |
|
1 (5.3) |
|
|
|
|
|
|
|
|
Five |
E |
CTR |
COT |
CTX |
AMC |
1 (5.3) |
E |
K |
S |
CTX |
AMC |
1 (7.1) |
|
Total MDR isolates |
19(51.4) |
|
14 (31.8) |
|||||||||
NB: AMC: Amoxyclav_Amoxicillin (Clavulanic Acid), CIP: Ciprofloxacin, C: Chloramphenicol, COT: Co-trimoxazole (Trimethoprim/Sulphamethoxazole), CTR: Ceftriaxone, CTX: Cefotaxime (Cefotaxime), DO: Doxycycline Hydrochloride, E: Erythromycin, GEN: Gentamicin, K: Kanamycin, NA: Nalidixic Acid, NIT: Nitrofurantoin, NX: Norfloxacin, S: Streptomycin, SF: Sulfisoxazole/Sulphamethoxazole and TR: Trimethoprim.
DISCUSSION
Bacterial contaminants of meat were assessed on meat specimens from the butchery houses. Meat contaminants (Salmonella species and E.coli) were detected in 39.8% (153/384) of the analyzed raw meat samples. Of which, 15.6% (n=60) and 20.8% (n=80) of the specimens were contaminated with Salmonella species and E.coli, respectively.
The overall contamination rate of Salmonella species in the butcher shop was 15.6%. This result was in line with prevalence reports from Ethiopia [27-29]. This was higher than the contamination rate reported from retail or butcher shops in Pakistan [30], Burkina Faso [31], and Ethiopia [16, 32-34]. But was lower than other reports from Ethiopia [29,35-38] and Mexico [39]. The overall contamination rate of E. coli in the butcher shop was 20.8%. This was consistent with other studies in the United States [40]. But higher than the other reports from Ethiopia [12,13,41]. However, it was lower than reports from Turkey [42], Canada [43], Burkina Faso [31], and Ethiopia [44- 47]. The variations in the prevalence of Salmonella species and E. coli might be due to the differences in meat handling for human consumption, personnel hygiene, and differences in hygiene measures taken during transportation. Other differences might be differences in sample type, differences in the origin of the samples or geographical differences, and differences in study methods and materials employed by the investigators.
Antimicrobial-resistant Salmonella species and E.coli isolates against antibiotics were presented. Salmonella isolates showed resistance to amoxyclav_amoxicillin (86.5%), erythromycin (56.8%), nitrofurantoin (43.2%), and streptomycin (29.7%). The resistance rate of nitrofurantoin in this study was in line with another study conducted in Ethiopia Amenu, 2012 [37]. Whereas E.coli isolates showed resistance to amoxyclav_ amoxicillin (88.6%), erythromycin (75%), cefotaxime (43.2%), streptomycin 43%), doxycycline hydrochloride (38.6%), and kanamycin 43.2%). The resistance rates of erythromycin [48] and streptomycin [12] conducted in Ethiopia were consistent with the present study. Also, streptomycin and cefotaxime which were carried out in Egypt were in line with our present finding [49].
The observed higher level of antimicrobial resistance might be attributed to the widespread use of antibiotics in animals for medication and other prophylaxis purposes. This difference might be due to the small sample sizes for the data, the nature of the drug, the presence of a different strain of the bacteria, and their low-frequency usage for prevention and control of disease in food animals in the study area. Antimicrobial resistance has the potential to adversely affect human health by causing illness that is more difficult to treat because of the resistance profile of the microorganism. This higher resistance profile of both isolates to amoxyclav_amoxicillin (clavulanic acid) and erythromycin might be attributed to a high level of utilization of this drug both in veterinary and human medicines due to its relatively cheaper price and ready availability to the local community in the current study area.
The overall multidrug resistance showed in Salmonella species was 51.4% whereas, for E. coli, the multidrug resistance observed was 31.8%. The multidrug resistance observed in this study showed resistance to three and above different classes of antibiotics [50]. Five (26.3%), two (10.5%), and one (5.3%) isolates of Salmonella species showed MDR to three and above, four and five classes of antibiotics respectively. Whereas five (35.7%), one (7.1%), and one (7.1%) isolates of E.coli showed MDR to three and above, four and five classes of antibiotics respectively. The MDR finding in the present study was in line with a report from Ethiopia [29]. However, the current finding result was higher than other studies conducted in Ethiopia [28,37]. On the other hand, our finding of multiple drug-resistance isolates of E.coli was lower than other reports from Ethiopia [40,47,48]. This difference in multi-drug resistance development in both Salmonella and E.coli might be due to the widespread and indiscriminate use of the commonly available antimicrobials both in veterinary and public health practices.
CONCLUSION
In the present study, 153 out of 384 (39.8%) of the samples were found to have a positive raw meat contamination rate. Poor hand washing regularly and not using gloves during meat handling showed significant association. Thirty-nine (88.6%) and thirty- three (75%) isolates of E.coli showed resistance to amoxiclav- amoxicillin and erythromycin respectively whereas thirty-two (86.5%) and twenty-one (56.8%) isolates of Salmonella species revealed resistance for amoxiclav-amoxicillin and erythromycin, respectively. Besides, the prevalence of multiple drug resistance such as 51.4% of Salmonella species and 31.8% of E.coli isolates.
There is a need to provide regular training to the butchery houses workers on best practices of food handling in all aspects of food hygiene and safety. Since the current study was conducted in a specific area, it is also recommended that further studies should be made using a larger sample size and covering a wider area.
ACKNOWLEDGMENTS
Special thanks to Mekelle University, College of Health Sciences, and Departments of Medical Microbiology and Immunology.
AUTHORS’ CONTRIBUTIONS
HAT, DG, AGK, and MA participated in designing the proposal, sample collection, experimental work, and writing and approving the manuscript. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was ethically approved by the ethical review committee of Mekelle University, College of Health Sciences. Permission was also obtained from the Tigray Health Bureau.
REFERENCES
- Foodborne Disease. A focus for Health Education. 2000.
- Jabbar MA, Grace D. Regulations for the safety of animal-source foods in selected Sub-Saharan African countries: Current status and their implications. Prepared for the Safe Food, Fair Food Project. In Livestock Research Inst. 2012.
- Pal M. Raw meat poses public health risks. Ethiop Herald. 2012; 2-3.
- Humphrey T, Jorgensen F. Pathogens on meat and infection in animals: Establishing a relationship using Campylobacter and Salmonella as examples. Meat Science. 2006; 74: 89-97.
- Pal M, Aton J, Paton A. Pathogenesis and diagnosis of Shiga toxin-producing Zoonosis. 2007; 100-134.
- Centers for Disease Control and Prevention. 2014.
- Batz MB, Hoffmann S, Morris JG. Ranking the disease burden of pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J Food Prot. 2012; 75: 1278-1291.
- European Centre for Disease Prevention and Control. The European Union Summary Report on Trends and Sources of Zoonotic Agents and Food-borne Outbreaks in 2012. EFSA J. 2014; 12: 35-47.
- The European Union summary report on trends and sources zoonosis, zoonotic agents and food-borne outbreaks in 2015. EFSAJ. 2016; 14: 34-46.
- Tadesse G, Gebremedhin EZ. Prevalence of Salmonella in raw animal products in Ethiopia: a meta-analysis. BMC Res Notes. 2015; 8: 163.
- Garedew KL, Wondafrash N, Feleke A. Identification of drug-resistant Salmonella from food handlers at the University of Gondar, Ethiopia. BMC Research Notes. 2014; 7: 545.
- Hiko A, Asrat D, Zewde G. Occurrence of Escherichia coli and Salmonella species in retail raw meat products in Ethiopia. J Infec Dev Ctries. 2008; 2: 389-393.
- Dulo F, Feleke A, Szonyi B, Fries R, Baumann MPO, Grace D. Isolation of multidrug-resistant Escherichia coli from goats in the Somali region of Ethiopia: a cross-sectional, abattoir-based study. PloS one. 2015; 10: 0142905.
- Ejeta G, Molla B, Alemayehu D, Muckle A. Salmonella serotypes isolated from minced meat beef, mutton and pork in Addis Ababa, Ethiopia. Revue Méd Vét. 2004; 155: 547-551.
- Mogessie A. Microbial flora and incidence of some foodborne pathogens on fresh raw beef from butcher s shops in Awassa, Ethiopia. Bull Anim Hlth Prod Afr. 1994; 42: 273-277.
- Tassew H, Abdissa A, Beyene G, Gebre-Selassie S. Microbial flora and foodborne pathogens on minced meat and their susceptibility to antimicrobial agents in Jimma Town, southwest Ethiopia. Ethiop J Health Sci. 2010; 20: 137- 143.
- Wendlandt S, Shen J, Kadlec K, Wang Y, Li B, Zhang W, et al. Multidrug resistance genes from animals that confer resistance to critically and highly important antimicrobial agents in human medicine. Trends Microbiol. 2015; 23: 44-54.
- Adams DA, Thomas KR, Jajosky RA, Foster L, Baroi G, Sharp P, et al. Summary of Notifiable Infectious Diseases and Conditions-United States, 2015. 2017.
- World Health Organization and European Union (WHO /EU). 2018.
- Ethiopia Antimicrobial Resistance Surveillance (EARS). Annual Report July 2017-August 2018 AMR-Report. 2018.
- Ministry of Agriculture (MOA). Meat transport and storage guidelines, for abattoirs and airport cargo terminal workers. Addis Ababa, Ethiopia. 2010.
- Bryant C. Investment opportunities in Mekelle, Tigray state, Ethiopia. 2016.
- Microbiology of Food and Animal Feeding Stuff-Horizontal Method for the Detection of Salmonella. 2002.
- El-Shamy HA, Bakr WI, Gomaa NF, Barheem OH. Evaluation of two enrichment broths, three plating media, and Elisa technique for the isolation of Salmonella from Dairy Products. J Egypt Public Health Assoc. 2008; 83: 133-145.
- Quinn P, Carter M, Markey B, Carter G. Clinical Veterinary Microbiology. Enterobacteriaceae. Wolfe Pub Spain, In. 2002; 209- 236.
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for anti-microbial susceptibility testing; Twenty-Second Informational Supplement. CLSI document. M100-S22. Wayne PA. 2017.
- Ejeta G, Molla B, Alemayehu D, Muckle A. Salmonella serotypes isolated from minced meat beef, mutton and pork in Addis Ababa, Ethiopia. Revue Méd. Vét. 2004; 155: 547-551.
- Ejo M, Garedew L, Alebachew Z, Worku W. Prevalence and Antimicrobial Resistance of Salmonella Isolated from Animal- Origin Food Items in Gondar, Ethiopia. Biomed Res Int. 2016; 2016: 4290506.
- Ashenafi M. Microbial flora and incidence of some foodborne pathogens on fresh beef from butcher’s shops in Awasa, Ethiopia. Bull Anim Health Prod Afr. 1994; 42: 273-277.
- Ali NH, Farooqui A, Khan A, Khan AY, Kazmi SU. Microbial contamination of raw meat and its environment in retail shops in Karachi. Pakistan. J Infect Dev Ctries. 2010; 4: 382-388.
- Kagambèga A, Haukka K, Siitonen A, Traoré AS, Barro N. Prevalence of Salmonella enterica and the Hygienic Indicator Escherichia coli in Raw Meat at Markets in Ouagadougou, Burkina Faso. J Food Protect. 2011; 74; 1547-1551.
- Molla B, Alemayehu D, Salah W. Sources and distribution of Salmonella serotypes isolated from food animals, slaughterhouse personnel and retail meat products in Ethiopia. Ethop J Helth Dev. 2003; 17: 63-70.
- Mengistu SH, Abayneh E, Shiferaw D. E.coli O157:H7 and Salmonella Species: Public Health Importance and Microbial Safety in Beef at Selected Slaughter Houses and Retail Shops in Eastern Ethiopia. J Vet Sci Technol. 2017; 8: 468.
- Zewdu E, Cornelius P. Antimicrobial resistance pattern of Salmonella serotypes isolated from food items and personnel in Addis Ababa, Ethiopia. Trop Anim Health Prod. 2009; 41: 241-249.
- Gebeya DN. Prevalence And Antibiotic Resistance of Salmonella Species In Meat Samples taken from a slaughterhouse and selected retail houses in Mekelle, Northern Ethiopia M.Sc. Thesis Haramaya University. 2011.
- Amenu A. Prevalence and Antibiotic Resistance of Salmonella Isolated From Beef in Arbaminch, Southern Ethiopia Haramaya University, MSc. Thesis. 2012.
- Garedew L, Hagos Z, Addis Z, Tesfaye R, Zegeye B. “Prevalence and antimicrobial susceptibility patterns of Salmonella isolates in association with hygienic status from butcher shops in Gondar town, Ethiopia’’. Antimicrob Resist Infect Control. 2015; 4: 21.
- Bhandare SG, Sherikarv AT, Paturkar AM, Waskar VS, Zende RJ. A comparison of microbial contamination of sheep/goat carcasses in a modern Indian abattoir and traditional meat shops. Food Contr. 2007; 18: 854-858.
- Zaidi MB, McDermott PF, Fedorka-Cray P, Leon V, Canche C, Hubert SK. Non-typhoidal Salmonella from human clinical cases, asymptomatic children, and raw retail meats in Yucatan, Mexico. Clin Infect Dis. 2006; 42: 21-28.
- Zhao C, Ge B, De Villena J, Sudler R, Yeh , Zhao S, et al. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the Greater Washington, D.C., area. Appl Environ Microbiol. 2001; 67: 5431-5436.
- Bekele T, Zewde G, Tefera G, Feleke A, Zerom K. Escherichia Coli in raw meat in Addis Ababa, Ethiopia: prevalence at an abattoir and retailers and antimicrobial susceptibility. Int J Food Contam. 2014; 1: 4.
- Siriken B. The microbiological quality of ground beef in Aydin and Afyon Provinces, Turkey. Revue Méd Vét. 2004; 155: 632-636.
- Doyle MP, Schoeni JL. Isolation of Escherichia coli from retail fresh meats and poultry. Appl Environ Microbiolol. 1987; 53: 2394-2396.
- FAO/WHO (Food and Agriculture Organization and World Health Organization FAO/WHO).Guidelines for slaughtering, meat cutting and further processing. 2012.
- Tassew H, Abdissa A, Beyene G, Gebre-Selassie S. Microbial flora and foodborne pathogens on minced meat and their susceptibility to antimicrobial agent. Ethiop J Helth Sci. 2010; 20: 137-143.
- Girma GM. Antibiogram of Escherichia coli strains disconnected from nourishment of bovine Origin in selected Woredas of Tigray, Ethiopia. Int J Bacteriol Res. 2015; 3: 148-153.
- Haileselassie M, Taddele H, Adhana K, Kalayou S. Food safety knowledge and practices of abattoir and butchery shops and the microbial profile of meat in Mekelle. Asian Pacific J Trop Biomed. 2013; 3: 407-409.
- Messele YE, Abdi RD, Shimels TY, Desiye TT, Bezina AE, Gebremeskel MW. Molecular determination of antimicrobial resistance in Escherichia coli isolated from raw meat in Addis Ababa and Bishoftu, Ethiopia. Ann Clin Microbiol Antimicrob. 2017; 16: 55.
- Moawad AA, Hotzel H, Awad O, Tomaso H, Neubauer H, Hafez HM, et al. Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 2017; 9: 57.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: An international expert proposal for interim standard dentitions for acquired resistance. Clin Microbiol Infect. 2012; 18: 268-281.