Nkx2-3 Homeobox Gene Plays an Essential Role in Later Stages of Orofacial Differentiation
- 1. Center for Regenerative and Developmental Biology, The Forsyth Institute, Cambridge, USA
- 2. Universitas Gadjah Mada, Yogyakarta, Indonesia
- 3. Cooper University Hospital, USA
- 4. Department of Surgery, Massachusetts General Hospital, USA
- 5. Department of Surgery, Harvard Medical School, USA
- 6. Shriners Hospital for Children, USA
- 7. Victor Chang Cardiac Research Institute, Darlinghurst, NSW, Australia; School of Clinical Medicine and School of Biotechnology and Biomolecular Science, Australia.
- #. Both the authors contributed equally
Abstract
The Nkx2-3 gene belongs to the NK2 class of homeobox genes that play important role in vertebrate development and share overlapping expression patterns. NK2s are involved in several pathways that lead to cell differentiation, migration, and maturation of the cells, indicating their essential role in the formation and homeostasis of the organism. Here we report that the Nkx2-3lacZΔHD/lacZΔHD mouse mutants have an orofacial phenotype affecting among other structures and the developing teeth. Specifically, we confirm that while incisors and upper molars are normal, the mandibular molars of the mutants have abnormal crown shape. Second, we provide for the first time evidence that the absence of Nkx2-3 affects the differentiation process of the two most essential dental cell populations, namely the ameloblasts and the odontoblasts. Macroscopic, histological, 3D micro-CT of the lower molars of Nkx2-3lacZDHD/lacZDHD mouse mutants exhibit enamel and dentin phenotypes with the expression of the Enamelin (Enam) and Dentin Sialophosphoprotein (DSPP) genes to be selectively reduced in Nkx2-3 deficient secretory ameloblasts and dentinoblasts, respectively.
KEYWORDS
- Nkx2-3 gene
- Vertebrate development
- Orofacial phenotype
- Differentiation
CITATION
Ruspita I, Das P, Kelangi S, Harvey RP, Bei M (2025) Nkx2-3 Homeobox Gene Plays an Essential Role in Later Stages of Orofacial Dif ferentiation. Pediatr Child Health 15(1): 1347.
INTRODUCTION
The Nkx2-3 gene belongs to the NK2 class of homeobox genes that play important role in vertebrate development and share overlapping expression patterns [1,2]. NK2s are involved in several pathways that lead to cell differentiation, migration, and maturation, indicating their essential role in the formation and homeostasis of the organism [1]. They are functionally distinct and have emerged as two different evolutionary branches [1,3]. The “neural” branch which is expressed in the central nervous system and the “cardiac” branch which is involved in patterning and differentiation of the heart and visceral mesoderm [1]. Members of both branches are also expressed in overlapping domains of the pharyngeal endoderm [1].
Nkx2-1 and Nkx2-8, for example, are responsible for lung epithelial development, thyroid and pituitary gland development [4]. In humans, the Nkx2-1 gene can cause neurological defects, congenital hypothyroidism, lung malformations, and is overexpressed in lung adenocarcinoma and small cell lung cancer [5-8]. Nkx2-2 and Nkx2-9 genes are necessary for the development of spinal cord interneurons and hindbrain visceral motor neurons [9]. The Nkx2-5, is responsible for early cardiac morphogenesis, angiogenesis, and hematopoiesis [10]. Its mutation causes an arrest in heart development and embryonic lethality in mice, while in humans, a single-nucleotide deletion mutation of NKX2-5, causes predominantly congenital atrial and ventricular septal defects and cardiac conduction abnormalities in adults [11,12]. In addition, Nkx2-5 gene plays critical role in lymphoid organ development and is required for spleen development [13,14].
Nkx2-3 deficiency in mice results in atrophic disorganized spleen, enlarged and disorganized colonic crypts, abnormal villus formation, and altered lymphocyte homing [15]. Several GWAS have shown an association between Nkx2-3 and inflammatory bowel diseases (IBDs). Single-nucleotide polymorphisms of Nkx2-3 (namely, rs10883365 and rs1190140) are associated with Crohn’s disease (CD) and Ulcerative Colitis (UC). Polymorphism rs10883365 may contribute to both CD and UC, whereas occurrence of the T allele of rs1190140 can increase the risk of CD [16]. Nkx2-3 also plays important role in normal developmental patterning and differentiation of several peripheral lymphoid organs and in hematopoietic/ lymphoid malignancies [15,17].
There are several reports indicating that Nkx2-3 participates in the development of other organs, as well. In mice, chick, Xenopus and zebrafish, nkx2-3 is known to be involved in the commitment and/or differentiation of cardiomyocytes [18]. It is also reported that it is involved in the development of several organs of head and neck, such as the thyroid, the sublingual salivary gland, the lingual epithelium, and the odontogenic epithelium [19]. Specifically, it has been reported that knockdown of Nkx2- 3 in mice leads to defects in the maturation and cellular organization of salivary glands and it is important for tooth cusp formation [19,20]. Mutations in the human homologue NKX2-3 are linked to developmental defects of the spleen and intestinal vasculature [21].
In this report, we first confirm previous studies where they showed that the Nkx2-3lacZΔHD/lacZΔHD mouse mutants have an orofacial phenotype affecting the developing teeth. Specifically, we confirm that while incisors and upper molars are normal, the mandibular molars of the mutants have abnormal crown shape. Second, we provide for the first time evidence that the absence of Nkx2-3 affects the differentiation process of the two most essential dental cell populations, namely the ameloblasts and the odontoblasts. Using 3D micro CT, histology and analysis by ISH of two representative markers-the Enam that is expressed exclusively by ameloblasts and the DSPP that is expressed exclusively by odontoblasts-we show that the ameloblasts and odontoblasts reach the secretory stage of their differentiation process affecting the amounts of enamel and dentin matrix deposited. In addition, we show that both pre-ameloblasts and pre-odontoblasts exhibit a polarization, adhesion and stratification defect, leading probably to a delay in the differentiation process.
MATERIALS AND METHODS
Mice and Genotyping
All animal studies and experimental procedures were conducted in accordance to the guidelines for the care and use of laboratory animals by the Massachusetts General Hospital, Boston, MA and the Forsyth Institute, Cambridge, MA. Nkx2-3lacZΔHD mice were maintained as both heterozygous and homozygous strains and were on a mixed 129SvJ x C57BL/6 background [22]. Embryos and postnatal pups (E18.5, P1,P3), were collected from wildtype and Nkx2-3 heterozygous. The day of plug discovery was designated as embryonic day 0.5 (E0.5). Age matched wildtype pups and/or embryos served as the appropriate controls.
Haematoxylin and Eosin Staining
For haematoxylin and eosin (H&E) staining, the mouse heads of embryonic day 18.5 (E18.5) and postnatal day 3 and 6 (P3 and P6) were fixed with 4% paraformaldehyde in PBS solution overnight and then were decalcified in 15% EDTA solution at 4°C for 0–7 days, depending on the age of the animals. The samples were processed for paraffin embedding, and serial sections of 5 μm in thickness were cut for H&E analyses.
ISH Assay
Embryonic Day 18.5 embryos and postnatal animals (P3) were collected and heads decapitated for making sagittal sections. The samples were immediately fixed in 4% paraformaldehyde. All samples were then dehydrated through graded ethanol series, embedded in paraffin, sectioned at 8 μm and processed for in situ hybridization (ISH), as previously described [23,24]. Sense and antisense primers were used to synthesize the probes in a PCR reaction. T7 primer sequence sites were added to the antisense sequence to generate the antisense probe by PCR method. The PCR products were gel purified (Qiagen Inc, Valencia, CA), labelled with DIG-UTP (Roche Biochemica, Mannhein, Germany), and used directly for hybridization. The sense probes was used as a negative control. The following are the sequences of probes used:
mNkx2-3-sense: (5’aagagacagcggcaggataa3’), mNkx2-3- antisense: (5’cttgggtctgtcctttctcg3”),
mEnam-sense: (5’ccagacttcctgcctcaaag3’),mEnam-antis ense:(5’aggactttcagtgggtgtgg3’),
mDSPP-sense: (5’aaacgaacaggggaacactg3’), mDSPP- antisense: (5’cctcactgctgttgtctc3’).
3D-Micro CT
Hemimandibles of Nkx2-3lacZΔHD/lacZΔHD and matched wild type littermate mice (2 months of age) were dissected and stored in 70% ethanol. Samples were analysed in ethanol using a MicroCT 40 system (Scanco Medical, Brüttisellen, Switzerland) at 70 kV, 114 μA, 8 watts, and 10 μm resolution with an integration time of 300 ms. Image sequences in dicom format were processed in ImageJ (National Institutes of Health) and resliced to achieve orientation of samples for comparison in images.
Statistical analysis
All experiments were performed independently at least three times (i.e., N=3) in triplicates, and when applicable, presented as an average ± standard error of the mean. Student t-test was used to determine p-values and P<0.05 was deemed to be significant.
FINDINGS/RESULTS
Enamel and dentin deposition is disturbed in the lower molars of NKX2-3 mutant adult mice.
Our macroscopic examination of teeth of Nkx2- 3lacZΔHD/lacZΔHD mutant mice (hereafter referred to as mutant) compared to WT counterparts at 2 months of age partially confirm previous reports that while the upper, lower incisors and upper molars appear normal, the mandibular molars of mutants are abnormal (data not shown) [19,22]. Indeed, all three mandibular molars erupt, and they are, some of the times, smaller in size [19,22]. Based on the previous reports, however, the defect of the two-month-old mutant mandibular molars was mainly the absence of cusps compared to their WT counterparts [19,22]. To better understand whether the cusps are indeed absent or whether they are formed, we examined the mineralized tissues of two-month-old (P60) wild type and mutant mandibular molars of mice, kept in soft diet, macroscopically and by 3D-microCT analysis (Figure 1). We show that the mandibular incisors’ shape, buccal and lingual surfaces are normal, as expected (Figure 1A),
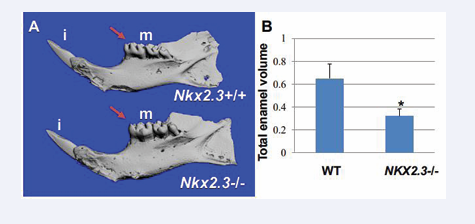
Figure 1: 3D-microCT analysis in the hemimandibles of Nkx2-3lacZΔHD/lacZΔHD mutant and matched wild type littermate mice (2 months of age).
(Figure 1A): the WT (top) and Nkx2-3-/- mutant (bottom) mandibular incisors’ (i) shape, buccal and lingual surfaces are normal. The cusps of the Nkx2-3-/- mutant mandibular molars (m) are formed but they are not sharp but rather smoother compared to WT counterparts (red arrows in Figure 1A). (Figure 1B): Bar graph presenting enamel volume of the mandibular teeth as mean ± s.e.m calculated from triplicates in each experimental group. There is a significant decrease of total enamel volume in Nkx2-3 mutants as compared to wild type controls (* P<0.05). Abbreviations: (i); incisors; (m): mandibular molars
with 100% penetrance. In contrast to previous reports, the cusps of the mandibular molars are formed and they are not sharp but rather smoother compared to WT counterparts (Figure 1A). To determine whether the defect in the occlusal surface of the crown of mutant mandibular molars is due to reduced enamel volume formation, we performed 3D-microCT in the hemimandibles of mutant and matched wild type littermate mice (2 months of age) and measured the enamel volume. We show that there is significant loss of enamel volume in the mutants compared to WT counterparts (Figure 1B), indicating that the differentiation process of ameloblasts is affected, “leading to weak enamel formation which becomes eroded most probably by mastication” [19]. Our results show that Nkx2-3 is not required for cusp morphogenesis, but it regulates the enamel deposition, suggesting an effect on the differentiation process of ameloblasts and/or odontoblasts.
The results of the 3D-microCT and that of our macroscopicanalyseswerefurtherconfirmedhistologically in E18. 5 embryos and post-natal day 3 (P3) mice (Figures 2A-D).
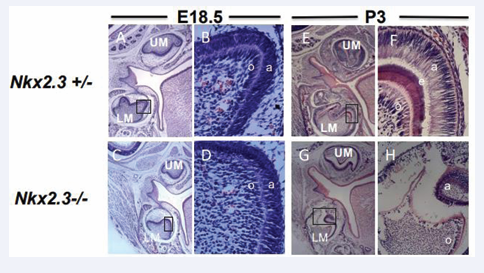
Figure 2: Differentiation defects in Nkx2-3 mutant teeth.
(A-D): H&E stained of E18.5 wild type and Nkx2-3 mutant first molar teeth, respectively. Insets in Figures. A and C are magnified in Figures. B and D. Both mutant pre-ameloblasts and odontoblasts are less polarized, and exhibit reduced cell-cell adhesion and stratification defects compared to WT controls. Abbreviations: UM: upper molar; LM: lower (mandibular) molar; O: odontoblasts; a: pre-ameloblasts. E-H): H&E stained of post- natal day 3 (P3) wild type and Nkx2-3 mutant first molar teeth, respectively. Insets in Figures E and G are magnified in Figures F and H. Both mutant ameloblasts and odontoblasts are less polarized, and exhibit reduced cell-cell adhesion and stratification defects compared to WT controls. Moreover, and in contrast to well-formed enamel and dentin matrix deposition, the mutant ameloblasts and odontoblasts are degenerating. Enamel (purple) and Dentin (red) are either hypoplastic or not formed. Abbreviations: UM: upper molar; LM: lower (mandibular) molar; o: odontoblasts; a: pre-ameloblasts. Scale: X50(A,C,G,E) and X400 (B,D,F,H) (N=3).
In control, WT and/or heterozygous mice (Figures 2 A, B), the pre-ameloblasts and pre-odontoblasts of the E18. 5 lower molars started their differentiation journey by being stratified and by beginning their polarization process prior reaching the pre-secretory and secretory stages (Figure 2A, B). In contrast, in the mutant E18. 5 embryos the cusps are formed but there is a delay in mutant lower molar pre-ameloblasts and pre-odontoblasts in their stratification and polarization process (Figure 2C, inset in 2D). Later in differentiation and during the early secretory stages at post-natal day 3 (P3) mice (Figure 2E- H), the mutant ameloblasts and odontoblasts gradually are less polarized compared to WT and/or heterozygous counterparts and their tight arrangement through stratification and adhesion was disturbed (Figures 2G,H), resulting in reduced amount of enamel and dentin formation (Figure 1D). Thus, the previous and latter results indicate that (i) the cusps are formed and (ii) that a later defect in differentiation affects the polarization and stratification processes of ameloblasts and odontoblasts- that are fundamental stages for normal volume enamel and tubular dentin formation-are disturbed in the Nkx2-3 mutant teeth leading to delay and/or abnormal deposition of both enamel and dentin matrices.
Enam and DSPP are under the control of Nkx2-3 transcription
To determine whether Nkx2-3 is associated with the defect in amelogenesis and dentinogenesis, in situ hybridization was performed in wild type mouse molar tooth germs at E.18.5, (Figure 3).
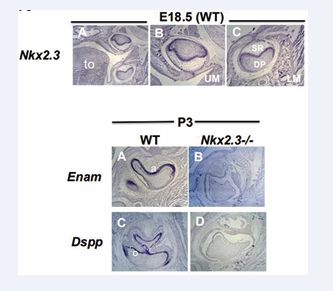
Figure 3: Nkx2-3 is essential for Enam gene expression during amelogenesis and DSPP gene expression during dentinogenesis:
Upper panel (Figures. 3A-C): ISH analyses of Nkx2-3 transcript in wild type E18.5: Nkx2-3 is expressed in stellate reticulum (SR), pre-ameloblasts, odontoblasts and dental papilla cells (DP).
Lower panel (Figures 3A-D): ISH analyses of Enam and DSPP transcripts in P3 wild type (A,C) and Nkx2-3 mutant (B, D) first lower molar teeth: Expression of Enam and DSPP is absent in Nkx2-3 mutant ameloblasts and odontoblasts, respectively, compared to wild type. Abbreviations: LM: lower molars; IN: incisors. Scale: X50 (upper panel A) and X100 (Upper panel B-C; lower panel A-D) (N=3).
Nkx2-3 is expressed by pre-secretory ameloblasts, odontoblasts, stellate reticulum and dental papilla cells (E18.5 (Figure 3 upper panel). Earlier reports refer to high Nkx2-3 expression in the region of the odontogenic ectoderm in the first pair of branchial arches at E10.5 and in the mandibular odontogenic epithelium at E13.5, but not in the maxillary one. Here we provide evidence that Nkx2-3 is also expressed in later stages of development during the cell differentiation processes of odontogenic derivatives. It is expressed in both, maxillary and mandibular molars in the epithelial-derived pre-secretory ameloblasts, stellate reticulum and in mesenchymal-derived dental papilla and odontoblasts.
To determine whether Nkx2-3 is required for ameloblast and odontoblast differentiation processes and whether this requirement is associated with the defect in amelogenesis and dentinogenesis, in situ hybridization was performed in wild type and Nkx2-3 mutant mouse lower molar tooth germs at postnatal day 3 (P3) for Enam and DSPP gene expression (Figure 3 lower panel).
Enam (Enamelin) is uniquely expressed by the ameloblasts and is expressed during the pre-secretory, secretory, transition and early maturation stages of ameloblast life cycle, while DSPP (Dentin Sialophosphoprotein) belongs to the small integrin- binding ligand N-linked glycoprotein (SIBLING) family of secreted phosphoproteins, which is expressed by the odontoblasts and are involved in bone and dentin deposition and mineralization [25-27].
A dramatic reduction is observed of Enam (Figure 3B) and DSPP (Figure 3D) expression in Nkx2-3 deficient molars (Figure 3B and Figure 3D), compared to wild type ones (Figure 3A and 3C) at P3, when ameloblasts and odontoblasts are at their pre-secretory and secretory stages, respectively.
Collectively, these results provide evidence for the first time that the absence of Nkx2-3 affects the differentiation process of the two most essential dental cell populations, namely the ameloblasts and the odontoblasts. In addition, we show that both pre-ameloblasts and pre-odontoblasts exhibit an early polarization, and stratification defect, leading probably to a delay in the differentiation process.
DISCUSSION
In this report, we confirm previous studies where they showed that the Nkx2-3 mutant mice have an orofacial phenotype affecting-among other organs-the morphogenesis of mandibular molars. Our 3D-microCT and histological studies show that while incisors and upper molars are normal, only the mutant mandibular molars have abnormal crown shape. Selective absence or selective phenotype of one tooth type versus another is observed in other mouse mutations. In Dlx1/Dlx2 homeobox double mutants, for example, only maxillary teeth are affected, while in activin βA mutant embryos the opposite is observed. The mandibular incisor and molar teeth fail to develop while the maxillary teeth are normal [28,29]. One plausible explanation for this selective phenotype is functional redundancy, where multiple transcription factors can perform similar functions, thus loss of one factor might be compensated by others. In the case of Nk2s, at the early stages of development, Nkx2-3 is co-expressed with the Nkx2-5 and Nkx2-6, suggesting complex regulatory interactions between members of this homeobox gene family and functional redundancy [30,31]. For example, the Nkx2-3, Nkx2-5 and Nkx2-6 appear to act redundantly in maintaining viability of early pharyngeal endodermal cells [30-32]. Whether other members of the NK2 family are co-expressed with Nkx2-3 in the oral ectoderm defining the maxillary dentition and lower incisors but not the mandibular molars, and whether these members compensate for the Nkx2-3 function needs further investigation.
In contrast to what it was reported in the past [19,20], however, we show using 3D-microCT and histological analysis that Nkx2-3 is not required for cusp morphogenesis and that the cusps of the mandibular molars are formed.
Biben and colleagues, who generated the Nkx2.3 mutants, suggested two potential roles for Nkx2-3. Either “Nkx2- 3 was necessary for the formation of enamel knots” and subsequently cusp morphogenesis, or that “terminal differentiation of ameloblasts is affected, leading to weak enamel which becomes eroded by mastication” [19]. To distinguish between the two possibilities, after weaning from their mothers, the mutant pups were kept in soft diet that could prevent severe enamel erosion giving the false impression of a flat-no cusp-crown appearance. Two months after their birth and in soft diet our 3D-microCT analysis show that the cusps are formed and that there is no significant difference in their width or height. What we observed, however, is that the occlusal surface of mutant mandibular molars is not as sharp but rather smoother compared to WT counterparts which is accompanied by a considerable decrease in the overall enamel volume. This suggests that the “terminal differentiation of ameloblasts is affected, leading to weak enamel which becomes eroded by mastication” [19]. Nkx2-3 ablation has also been analysed for effects on early dental development using ex vivo cultures of E13.5 molars by Han and colleagues [20]. Specifically, E13.5 mandibular tooth germs were dissected and then transfected with control or Nkx2-3 siRNA and after 1 week of culture, tooth cusp sizes in both groups were measured [20]. Based on this experimental approach, the authors claimed that the cusps’ height in the presence of Nkx2-3 siRNA was significantly reduced as compared with the control, suggesting that Nkx2-3 is critical for molar cusp formation [20]. Our in vivo experiments do not support this finding. As potential explanation for this discrepancy is the fact that in the ex vivo experiments the authors did not use chemically defined medium where all the chemical components are precisely known, allowing thus for a more controlled environment when studying an organ removed from a living organism outside the body.
On the other hand, Han and colleagues using their ex vivo organ culture, they showed that Nkx2-3 is expressed in the stellate reticulum (SR) and dental papilla (DP) cells by immunohistochemistry. Our ISH experiments in E18.5 WT embryos show that Nkx2-3 gene is indeed expressed in SR, DP but also in pre-ameloblasts and odontoblasts, partially confirming their findings. Han and colleagues also show that Nkx2-3 regulates dental epithelial cell proliferation by inducing p21 [20]. In E13.5 mandibular tooth germs transfected with control or Nkx2-3 siRNA, and cultured for 7 days the number of Ki6-positive cells were increased in the presence of Nkx2-3 siRNA in the stellate reticulum and dental papilla cells [20]. The stellate reticulum is a very particular type of star-shaped cells in the enamel organ and recent studies have shown that plays a crucial role in establishing the polarity of ameloblasts and by providing guidance to the ameloblasts to properly secrete enamel in the right direction [33-37]. An abnormally increased or reduced SR cell population affects the process of polarization and that of proper enamel deposition by ameloblasts [33-37]. In that context, our findings of (i) strong expression of Nkx2-3 in the SR and pre-ameloblasts of E18.5 WT molar teeth, by ISH (ii) the polarization, adhesion and stratification defect of Nkx2-3 mutant molar ameloblasts in E18.5 and P3, by histology (iii) the reduced enamel volume by 3D-microCT, and (iv) the dramatic downregulation of an ameloblast differentiation-specific marker, that of Enam gene, in the Nkx2-3 mutant mandibular molars, by ISH, provide a link between the role of Nkx2-3 in the SR, polarization, adhesion and stratification defect of ameloblasts, leading probably to a delay in the differentiation process and proper enamel deposition. It seems that the mutant ameloblasts reach the secretory stage of their differentiation process but the amount or the timing of enamel matrix is not deposited properly. Detailed analysis with polarization, adhesion and differentiation markers is needed that will further our understanding of the role of Nkx2-3 gene in ameloblast cell differentiation process. Interestingly other transcription factors such as the Ctip2/Bcl11b, SATB1, Msx2, Epiprofin are also expressed in the stellate reticulum and when they are mutated in mice, the ameloblasts exhibit, polarization, adhesion and enamel matrix deposition defects [33-35]. These results suggest that Nkx2-3 may also function as part of a critical regulatory network of transcription factors that determine epithelial cell fate and differentiation during tooth morphogenesis.
Another interesting finding is the odontoblast phenotype in the mutant molars. We show that Nkx2-3 is expressed by the mesenchymal-derived dental papilla cells and the odontoblasts. We also show that there is mild polarization and adhesion defect in the odontoblasts by histology and reduced DSPP expression in the Nkx2- 3 mutant odontoblasts suggesting a role for Nkx2-3 in the normal differentiation and function of odontoblasts. Although, further analysis with polarization, adhesion and differentiation markers is needed to help our understanding of the role of Nkx2-3 gene in odontoblast cell differentiation process, the lack of an obvious dentin phenotype at 2 months after birth is indicative of a later compensation mechanism, or that the role of Nkx2-3 in odontoblast differentiation process is temporary. Given that the Nkx2-3 transcription factor is expressed in many mesenchymal or epithelial-derived cells, beyond ameloblasts or odontoblasts, and that the Nkx2-3 mutant has phenotypes in other organs as well, the severity of a phenotype or how indirect effects might control differentiation could be an issue. To circumvent this potential limitation, future studies with conditional Nkx2.3 mutant mice could be valuable tools to study amelogenesis and dentinogenesis specifically at different stages of differentiation and homeostasis.
In conclusion, thekeyroleof Nkx2-3 in the differentiation, fate and commitment of different cell types is well known [18-20]. In mice, chick, Xenopus and zebrafish, Nkx2-3 is involved in the commitment, maturation, cellular organization and overall differentiation process of cardiomyocytes, of sublingual salivary gland, thyroid and lingual epithelial cells [18-20]. Here, we report for the first time that the absence of Nkx2.3 affects the differentiation process of the two most essential dental cell populations, namely the ameloblasts and the odontoblasts. In addition, we show that both pre-ameloblasts and pre-odontoblasts exhibit an early polarization, and stratification defect, leading probably to a delay in the differentiation process. Our findings corroborate and add to the overall concept that the NK2s have specific overlapping or distinct expression patterns creating a special transcriptional setting, which is essential in development, differentiation, and adult tissue patterning in an organ-, tissue- and cell-specific manner.
Data Availability Statement
All datasets generated for this study are included in the article.
Ethics Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of Forsyth Institute and Massachusetts General Hospital, Boston, MA (2013N000213, Date: 12/22/2023).
AUTHOR CONTRIBUTIONS
MB contributed to the concept and design of the study. IR and MB contributed to the acquisition of data. IR, PD, and MB prepared the figures. IR, PD, and MB contributed to the data analysis and interpretation. IR, PD and MB wrote the manuscript. IR, PD, SK, RH, and MB contributed to the editing and the critical revision for intellectual content. All authors have approved the final version of the submitted manuscript.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
FUNDING
The study was supported by funds from NIH [grant, R21DE028091] and the MGH Executive Committee Of Research to M.B. RPH was funded by grants from the National Health and Medical Research Council of Australia (NHMRC; 354400, 2008743).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- Harvey RP. NK-2 Homeobox Genes and Heart Development. Dev Biol.1996; 178: 203-216.
- McMahon A. Neural patterning: The role of Nkx genes in the ventral spinal cord. Genes Dev. 2000; 14: 2261-2264.
- Hombria JC, Lovegrove B. Beyond homeosis-HOX function in morphogenesis and organogenesis. Differentiation. 2003; 71: 461-476.
- Kimura S, Hara Y, Pineau T, Fernandez salguera P, Fox CH, Ward JM, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996; 10: 60-69.
- Safi KH, Bernat JA, Keegan CE, Ahmad A, Hershenson MB, Arteta M. Interstitial lung disease of infancy caused by a new NKX2-1 mutation. Clin Case Rep. 2017; 5: 739-743.
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007; 450: 893-898.
- Yang L, Lin M, Ruan WJ, Dong LL, Chen EG, Wu Xh, et al. Nkx2-1: a novel tumor biomarker of lung cancer. J Zhejiang Univ Sci B. 2012; 13: 855-866.
- Hsu DS, Acharya CR, Balakumaran BS, Riedel RF, Kim MK, Stevenson M, et al. Characterizing the developmental pathways TTF-1, NKX2-8, and PAX9 in lung cancer. Proc Nat Acad Sci U S A. 2009; 106: 5312- 5317.
- Briscoe J, Sussel L, Serup P, Hartigan O Connor P, Jessell TM, et al. Homeobox gene Nkx2.2 and specification of neuronal identity by graded sonic hedgehog signalling. Nature. 1999; 398: 622-627.
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995; 9: 1654-666.
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999; 126: 1269-1280.
- Ellesoe SG, Johansen MM, Bjerre JV, Hjortdal VE, Brunak S, Larsen LA. Familial atrial septal defect and sudden cardiac death: identification of a novel NKX2-5 mutation and a review of the literature. Congenit Heart Dis. 2016; 11: 283-290.
- Tanaka M, Yamasaki N, Izumo S. Phenotypic characterization of the murine Nkx2.6 homeobox gene by gene targeting. Mol Cell Biol. 2000; 20: 2874-2879.
- Lettice LA, Purdie LA, Carlson GJ, Kilanowski F, Dorin J, Hill RE. The mouse bagpipe gene controls development of axial skeleton, skull, and spleen. Proc Nat Acad Sci USA. 1999; 96: 9695-9700.
- Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999; 126: 2215-2225
- Lu X, Tang L, Li K, Zheng JY, Zhao P, Tao Y, et al. Contribution of NKX2–3 polymorphisms to inflammatory Bowel diseases: a meta- analysis of 35358 subjects. Sci Rep. 2014; 4: 3924
- Vojkovics D, Kellermayer Z, Kajtár B, Roncador G, Vincze Á, Balogh P. Nkx2-3-A slippery slope from development through inflammation toward hematopoietic malignancies Biomark. Insights. 2018; 13.
- Lyons GE. Vertebrate heart development Curr Opin Genet Dev. 1996; 6: 454-460
- Biben C, Wang CC, Harvey RP. NK-2 class homeobox genes and pharyngeal/oral patterning: nkx2-3 is required for salivary gland and tooth morphogenesis. Int J Dev Biol. 2002; 46: 415-422
- Han X, Yoshizaki K, Miyazaki K, Arai C, Funada K, Yuta T, et al. The transcription factor NKX2-3 mediates p21 expression and ectodysplasin-A signaling in the enamel knot for cusp formation in tooth development. J Biol Chem. 2018; 293:14572-14584
- Kerkhofs C, Stevens SJ, Faust SN, Rae W, Williams AP, Wurm P, et al. Mutations in RPSA and NKX2-3 link development of the spleen and intestinal vasculature. Hum Mutat. 2020; 41: 196-202.
- Wang C, Biben C, Robb L, Nassir F, Barnett C, Davidson NO, et al. Homeodomain Factor Nkx2-3 Controls Regional Expression of Leukocyte Homing CoreceptorMAdCAM-1 in Specialised Endothelial Cells of the Viscera Dev Biol. 2001; 224: 152-167.
- Ruspita I, Das P, Xia Y, Kelangi S, Miyoshi K, Noma T, et al. An Msx2- Sp6-Follistatin Pathway Operates During Late Stages of Tooth Development to Control Amelogenesis. Front Physiol. 2020; 11: 582610.
- Ruspita I, Das P, Miyoshi K, Snead ML, Noma T, Bei M. Enam expression is regulated by Msx2. Dev Dyn. 2023; 252: 1292-1302
- Daubert DM, Kelley JL, Udod YG, Habor C, Kleist CG, Furman IK, et al. Human enamel thickness and ENAM polymorphism. Int J Oral Sci. 2016; 8: 93-97.
- Papagerakis P, Hu Y, Ye L, Feng JQ, Simmer JP, Hu JC. Identifying promoter elements necessary for enamelin tissue-specific expression. Cells Tissues Organs. 2009; 189: 98-104.
- MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4: dentin phosphoprotein DNA sequence determination. J Biol Chem. 1997; 272: 835-842.
- Qiu M, Bulforne A, Ghattas I, Meneses JJ, Chistensen I, Sharpe PT, et al. Role of Dlx Homeobox Genes in Proximodistal Patterning of the Branchial Arches: Mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 Alter Morphogenesis of Proximal Skeletal and Soft Tissue Structures Derived from the First and Second Arches. Dev Biol. 1997; 185: 165- 184.
- Ferguson CA, Tucker AS, Christensen L, Lau AL, Matzuk MM, Sharpe PT. Activin is an essential early mesenchymal signal in tooth development that is required for patterning of the murine dentition. Genes Dev. 1998; 12: 2636-2649.
- Tanaka M, Schinke M, Liao HS, Yamasaki N, Izumo S. Nkx2.5 and Nkx2.6, homologs of drosophila tinman, are required for developmentof the pharynx. Mol Cell Biol. 2001; 21: 4391-4398.
- Tanaka M, Yamasaki N, Izumo S. Phenotypic characterisation of the murine Nkx2.6 homeobox gene by gene targeting. Mol Cell Biol. 2000; 20: 2874-2879.
- Trainor PA, Krumlauf R. Hox genes, neural crest cells and branchial arch patterning. Curr Opin Cell biol. 2001; 13: 698-705.
- Golonzhka O, Metzger D, Bornert JM, Bay BK, Gross MK, Kioussi C, et al. Ctip2/Bcl11b controls ameloblast formation during mammalian odontogenesis. Proc Natl Acad Sci U S A. 2009; 106: 4278-4283.
- Zhang Y, Zheng L, Le M, Nakano Y, Chan B, Huang Y, et al. SATB1 establishes ameloblast cell polarity and regulates directional amelogenin secretion for enamel formation. BMC Biol. 2019; 17: 104.
- Bei M*, Stowell S, Maas R. Msx2 controls ameloblast terminal differentiation. Dev Dyn. 2004; 231: 758-765.