The Amazing Multi-Valency of the Hsp70 Chaperones
- 1. Department of Biological Chemistry, The University of Michigan Medical School, USA
- 2. Institute for Neurodegenerative Disease, University of California at San Francisco, USA
Abstract
Hsp70 proteins are keys to maintaining intra-cellular protein homeostasis. To carry out this task, they employ a large number of co-chaperones and adapter proteins. Here we review what is known about the interaction between the chaperones and partners, with a strong slant towards structural biology. Hsp70s in general and Hsc70 (HSPA8) in particular, display an amazing array of interfaces with their protein co-factors. We also reviewed the known interactions between Hsp70s and active compounds that may become leads towards Hsp70 modulation for treatment of a variety of diseases.
Keywords
Hsp70 proteins; Protein chaperones.
CITATION
Zuiderweg ERP, Gestwicki JE (2016) The Amazing Multi-Valency of the Hsp70 Chaperones. JSM Cell Dev Biol 4(1): 1019.
INTRODUCTION
Hsp70 chaperones are highly conserved in all kingdoms; in animals, they are an important member of the collection of protein chaperones including Hsp60, Hsp70, Hsp90 and small Hsps [1]. The archetypical Hsp70 is called DnaK in bacteria and it functions in protein trafficking and protein refolding cycles, acting together with a nucleotide exchange factor (NEF), termed GrpE, and a J-protein, called DnaJ. In the presence of ATP, this trio is capable of refolding denatured luciferase in vitro, and can thus be classified as a protein refolding machinery [2]. While the prokaryotic system has a single Hsp70, NEF and J protein, the number of genes has expanded in eukaryotes. For example, the yeast genome codes for 14 Hsp70s, 20 J-proteins [3] and 3 NEFs [4]. In humans, there are 13 Hsp70s, 50 J-proteins and eight NEFs [5]. The original protein refolding function of the Hsp70/J/ NEF trio seems to be maintained throughout the kingdoms, but eukaryotes require more isoforms to enable the system to have representatives in each organelle (e.g. ER, mitochondria, cytoplasm/nucleus). Moreover, the expansion of the co chaperones seems to have allowed the system to adapt to a wider range of specialized functions, such as trafficking and signaling. Finally, higher eukaryotes have also added new partners for the Hsp70s, including HOP, HIP, CHIP and other proteins, that couple it to additional cellular functionality, such as proteosomal and autophagosomal degradation system and to the Hsp90 protein folding machinery. A key to understanding Hsp70 biology, especially in eukaryotes, is to understand how it coordinates with an expanded number of co-chaperones to enable its broad functions.
HSPA8 (Hsc70; constitutive) and HSPA1 (Hsp70-1; inducible) are the two major human Hsp70s of the cytoplasm and nucleus. Broadly speaking, these proteins are involved in four general activities in the cell: (i) binding and release of native and misfolded proteins to favor protein (re)folding cycles [6]; (ii) transporting unfolded proteins through membranes to enable delivery of cargo to organelles [7]; (iii) recruiting proteins to the proteasome for turnover [8] and (iv) bringing proteins to the endosome/lysosome for chaperone-mediated autophagy [9]. These diverse functions are achieved by Hsc70 interactions with different molecular partners: J proteins, NEFs and Hsp90 for function (i); membrane transporting systems for role (ii), interaction with ubiquitin ligases and NEFs for role (iii) and interactions with phosphoserine lipids and LAMP-2A for role (iv). Even from this simplified overview of Hsp70, it is clear that co-chaperones and other partners are the keys to understanding function. Another lesson is that each of the categories of chaperone function involves multiple components.
Because of these broad functions, theHsp70s are increasingly being seen as drug targets across a range of diseases [10]. For example, HSPA1 expression is induced in cancer cells, where it inhibits apoptosis. The levels of HSPA1 are even further elevated in response to chemotherapy, which may partially limit the effectiveness of Hsp90 inhibitor treatments. In another example, HSPA8 enhances the lifetime of aberrant tau protein in neurons, likely contributing to neurodegenerative disease. Hence, in addition to the basic scientific interest in delineating the Hsp70 functional cycles, there are translational implications in better understanding these mechanisms. How might we selectively disrupt some disease-associated functions of Hsp70 and not other, housekeeping activities? Several synthetic modulators of Hsp70 have recently emerged that have begun to answer this question.
In this review, we will focus on the structural and biophysical aspects of how Hsp70s bind to their different partners, as revealed by X-ray crystallography and multi-dimensional NMR spectroscopy. Ideally, one would like to focus on the Hsp70s of one species, and preferentially humans. However, much of the early structural biology was focused on the E.coli system of DnaK/ GrpE/DnaJ. Herein we want to cast a wide net, so we will assume that the level of homology between the different Hsp70 proteins and co-factors is sufficiently high so that structural knowledge can be safely extrapolated between orthologs and paralogs. For example, Hsc70 is 80% homologous and 55% identical to DnaK. In this review, we will use the name Hsp70 if we do not want to discriminate between homologues, and will use the systematic names as proposed by [5] if we do.
THE ATPASE CYCLE OF HSP70S
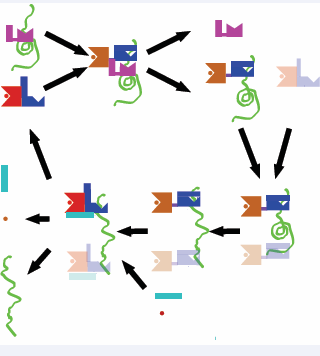
The ATP hydrolysis cycle of DnaK is illustrated in Figure (1).
Figure 1: Hsp70 functional cycle.The Hsp70 substrate unfolding cycle. Red and Blue: Hsp70-ATP/Apo state; Brown and Blue: Hsp70 ADP/SUB state; Pink: DnaJ; Cyan: NEF; Green: Substrate. Red dot: ATP; Brown dot: ADP. The transparent Hsp70 shapes represent hypothetical additional substrate binding.
The Hsp70 chaperones consist of a Nucleotide Binding Domain (NBD), Substrate Binding Domain (SBD) and a LID. With the LID open, the exposed hydrophobic cleft in the SBD binds to the exposed hydrophobic regions of misfolded / unfolded proteins with a (weak) affinity of approximately 10 µM. The co chaperone DnaJ helps to stabilize this interaction by also binding to unfolded areas of the same substrate, and by stimulating, with its 70-residue J-domain, ATP hydrolysis in the NBD. DnaK, now in the ADP-state, closes its LID and binds the substrate with a 100-to 1000-fold tighter affinity. Whether DnaJ dissociates at this stage of the cycle or later, is currently unknown. A process of entropic pulling, potentially involving several Hsp70s, progressively unfolds the substrate[11], which remains bound. Subsequently, a NEF enables ADP ATP exchange, with a concomitant decrease in the affinity of Hsp70 for its substrate. Possibly aided by direct competition of the NEF with the bound substrate[12] the substrate is now released in a folding-competent form, either to the cytosol or to GroEL for refolding.
The human Hsp70 system conforms to this basic ATPase cycle, although there are differences in complexity. As mentioned above, the human genome codes for 13 Hsp70 isoforms [5]. The Hsp70s are highly homologous and consist of three constitutively expressed proteins, Hsc70 (HSPA8) in the cytosol and nucleus, Bip (HSPA5) in the endoplasmic reticulum and Mortalin (HSPA9) in the mitochondria. The expression of the other Hsp70s, especially Hsp70-1 (HSPA1a/b), is induced upon cellular stress. These stress-induced factors appear to be dedicated to maintaining cellular proteostasis. For protein refolding cycles the human Hsp70s utilize 50 different J-proteins and at least 8 different NEFs. The J-proteins are characterized by the inclusion of a conserved J-domain. Some of them (HDJA1 and HDJA2) are homologous with the bacterial DnaJ even outside the J-domain, while others have completely unrelated motifs outside the J-domain. It is believed that these different J-proteins can bestow tissue- and functional specificity to the otherwise “generalist” Hsp70 family [13]. The human NEF family contains 6 BAG proteins (Bcl-2-associated AnthanoGene), aHsp110 protein, HSPBP1, and HME. HME is restricted to the mitochondria and shows homology to E. coli GrpE. The other NEFs are quite different from the bacterial prototype and share no obvious sequence homology. The Hsp110’s are structurally related to the Hsp70s themselves, using its NBD to act as a NEF [14,15]. The BAG proteins use a conserved BAG domain to interact with the top lobes of the NBD to favor nucleotide cycling [16]. HspBP1 has an α-helical fold containing four armadillo-like repeats[17]. Regardless of the exact identity of theJ-protein orNEF, each partner is thought to regulate the human Hsp70protein refolding machinery in analogous fashion as the DnaK/DnaJ/GrpE cycle [18]. At the end of the cycle,HSPA1 or HSPA8 can transfer the substrate to Hsp90, either directly or in a complex with HOP (Hsp organizing protein) [18]. HOP contains two tetratricopeptide repeats (termed TPR1 and TPR2A), which bind to EEVD-COOH motifs at the C-terminal tails of Hsp70 and Hsp90, respectively. Unfolded Hsp70 substrate is also being transferred to TRIC, the mammalian GroEL homologue, for refolding [19].
Why does the human Hsp70 system have so many co chaperones? We use the BAG family of NEFs to illustrate the current ideas. The six BAG proteins contain an alpha-helical domain of approximately 100 amino acids called BAG domain, which conveys the NEF function. However, BAG proteins have other functionalities as well. All of the BAG-family proteins function as adapter proteins forming complexes with different signaling molecules and the Hsp70’s. For instance, BAG-1 has besides its BAG domain a TXEEX domain involved in DNA binding and transcription activation and an Ubiquitin-like domain, which can target BAG1-Hsp70 complexes to the proteasome for degradation of specific client proteins.BAG3 is one of the largest BAG proteins and contains multiple protein-protein interaction motifs: a WW domain, multiple PXXP motifs, and two IPV motifs, allowing for PPxY protein binding, SH3 protein binding, and small heat shock protein binding respectively [20]. The non BAG domains of BAG3 have recently been shown to interact with HSPA8 SBD. The BAG3-HSPA8 complex stabilizes several oncogenes and is a particularly interesting drug target [21]. BAG1 and BAG3 represent key players of cellular by stimulating the turnover of polyubiquitinated proteins by proteasomal and autophagic degradation pathways, respectively [22]. BAG-4, containing a single BAG domain and another 350 residues for which no domain annotation is known, is a regulator preventing constitutive signaling by death domain receptors [23]. BAG 5consists four BAG domains[24]. Currently not much is known about the role of BAG-5 in cells other that it binds to Hsp70. Bag 6 has been called a “Jack of all trades”[25], as it interacts with nucleoprotein p300 in response to DNA damage andinteracts with histone methyltransferases. BAG-6 is required for the accumulation of HSPA1upon heat shock while HSPA1leads to the proteosomal degradation of BAG-6. Thus, all of the BAG proteins enhance nucleotide exchange in Hsp70 through their BAG domains, yet they engender dramatically different functionality by the remainder of their sequences. In other words, depending on which of the BAGs 1-6 is engaged, the fate of the Hsp70-bound protein might be quite different.
On top of the diversity of the BAG proteins, these proteins are all antagonized by the Hsp Interacting Protein (HIP), a homodimer containing several TPR domains [26]. HIP strongly stabilizes the Hsp70 ADP state, but may have also additional functions through its TPR domains. It was shown that HSPA1, HDJ1 and HIP, without a NEF, can promote protein refolding. Thus, the story of Hsp70 function seems to be one in which co chaperones and other partners combine with Hsp70 in dynamic ways, creating multi-valent complexes that dictate outcomes.
HSPA8 shuttles proteins to the cellular degradation pathways
Another important aspect of Hsp70 function is that it bridges misfolded proteins (or “clients”) to distinctcellular fates. To illustrate this point, we briefly review how Hsp70s can direct clients to either the ubiquitin-proteasome system (UPS) or the chaperone-mediated autophagy (CMA) pathway. Indeed, it is because of Hsp70’s interactions with these divergent paths that it is sometimes called the “triage” chaperone [27]. In short, it interacts with unfolded proteins and helps determine whether it should be folded (and saved) or whether it is hopelessly misfolded (and should be removed). The chaperone also seems to determine which of the two major degradation pathways should be employed. How does Hsp70 do this for thousands of different polypeptide sequences and countless different protein folds?
One of the best-understood roles of HSPA8 is in proteosomal degradation. The C-terminal of Hsp70 Interacting Protein (CHIP)contains a TPR domain, whichbinds to the C-terminal IEEVD motif of HSPA1 and HSPA8 [28]. CHIP also contains an Ubox domain, which binds to ubiquitin ligases such as UBCH5. The latter ubiquininates the HSPA8-bound substrate (and HSPA8 itself), which targets the complex to the proteasome for degradation. Thus, CHIP links HSPA8 to the UPS and helps regulate protein turnover. Surprisingly, the same chaperone also plays a crucial role in Chaperone Mediated Autophagy (CMA) [29] and endosomal microautophagy[30]. In CMA, HSPA8 recognizes a subset of cytosolic proteins containing a KFERQ motif [31]. The HSPA8-substrate complex docks to the lysosomal receptor LAMP2A after which substrate is translocated into the lysosome. For Endosomal microautophagy HSPA8 also interacts with KFERQ-type proteins for ESCRT I and III mediated delivery into an endosome. Despite progress in understanding the players in these degradation pathways, the molecular mechanisms are mysterious. One of the most important, open questions is whether HSPA8 uses other, dedicated co-chaperones to help make these triage decisions. In theory, assembly of a discrete collection of co-chaperones, perhaps directed by structural features of the misfolded protein client, could be sufficient to explain how “decisions: are made. In other words, protein-protein interactions and molecular recognition by multiple chaperone surfaces might be the key.
Hsp70 internal interfaces: the differences between the ATP and ADP states
Hsp70s have an NBD, SBD, LID and tail, connected by linkers in that order. The Hsp70 chaperone structural biology field is replete of crystal and solution structures of the individual NBDs (first: HSPA8 [32]) and SBDs (first: DnaK [33]) of Hsp70 orthologs and paralogs, but has been plagued by a paucity of valid structures for the full-length proteins, which became available since 2009 for DnaK. No structures for full-length mammalian Hsp70s are available as of yet. And, even today, no comparison between ATP and ADP states for any Hsp70 can be made solely on basis of experimental data.
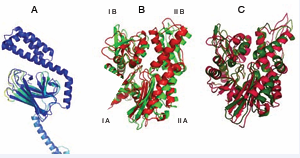
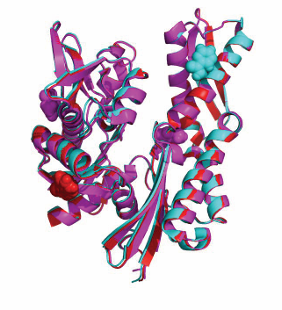
The first structure of a full-length Hsp70, DnaK(1-605), containing NBD, SBD and LID [34] was based on the crystal structures of the isolated SBD bound to the peptide NRLLLTG (1DKX) and the isolated NBD of DnaK without nucleotide and bound to GrpE (1DKG). NMR data provided the relative orientation and mobility range of these domains. A key finding was that the NBD and SBD domains were free to move with respect to each other in this state like “beads on a string” (Figure 2).
Figure 2: Structures of the E.coli Hsp70 protein DnaK in the ADP/ NRLLLTG state (top, 2KHO) and in the ATP/Apo state (bottom, 4B9Q). The NBDs (green) have the same orientations. Red: beta-domain of the SBD; Blue: Lid domain of the SBD. DnaK contains another 30 unstructured residues at the C-terminus of the lid (not shown).
Structures of the ATP state of NBD-SBD constructs of E.coli DnaK, without peptide, were determined by two groups [35,36]. These crystal structures show the SBD firmly docked to the NBD, and the alpha helical SBD LID domain docked to the NBD instead of the SBD (Figure 2). NMR data indicated that the NBD-SBD docking in the ATP state occurs in solution as well [37,38]. The enormous differences in inter-domain docking between the ATP- and ADP bound states have been discussed and evaluated in detail in the original papers [35,36] and several reviews since, and will not be repeated here, except to point out that Hsp70 exposes and covers large and different inter-domain surfaces in the two states.
The DnaK SBD in the peptide-bound state is very well ordered[33], while the apo state is disordered around the substrate binding cleft [35,36]. This is illustrated in Figure (3A), where the SBD is colored according to the crystallographic B-factor. High B-factors indicate disorder, which can indicate either (frozen out) dynamics or static disorder. The figure thus shows that the loops surrounding the substrate are highly disordered in the apo state. Early NMR data of DnaK SBD indicated that this disorder is based on dynamics at the micro-second time scale [37-39].The structural and dynamical differences between DnaK in the apo and peptide-bound state have been further studied in solution by Zhuravleva et al. [40].
There is no direct experimental comparison possible for the DnaK NBD in the ATP and ADP state, by account that no structure has been determined for the latter. So, what happens to the NBD between ATP and ADP states? Many crystal structures are available for the mammalian Hsp70NBDs in the ADP state and ATP (analogue) states. Regretfully, these structures are all identical within experimental precision, providing no insight into ATP/ADP-induced differences that must occur. The best we can do for now, is to construct a homology model of HSPA8 NBD in the ATP state, based on the crystal structure of DnaK in the ATP state, and compare it with the crystal structure of HSPA8 NBD in the ADP state (Figure 3B).
Figure 3 A: Overlay of the SBD of DnaK E.coli in the NRLLLTG (1DKX) and apo (4B9Q) state. The figure ribbon is colored according to the B-factor ranging from blue (high order) to yellow (low order). In this representation, the NRLLLTG state is dark blue throughout. B: Overlay of the NBD of HSPA8 in the ADP state (green, 3HSC), with a model of HSPA8 NBD based on DnaK in the ATP state (red, 4B9Q). The subdomain nomenclature is shown. C: Model of the ADP state of the NBD of DnaK from Thermus thermophilus (green, 4B9Q) based on DnaK E. coli, and the experimental rotational changes occurring upon binding of AMPPNP state (red).
The model and structure are superposed on subdomain IA (left bottom in figure). According to this model, the left and right hand lobes of the human HSPA8 NBD rotate by 10 degrees between the states. Furthermore, the cleft between lobes I and II is compressed in the ATP state and mayor changes occur in domain IB (left top).This model likely has some validity: early NMR work indicated that ATP/ADP binding causes major conformational changes in the HSPA8 NBD [41]. Furthermore, Bhattacharya [42]showed that the left and right hand lobes of the NBD of DnaK Thermus thermophilus rotate by 10 degrees between ADP and ATP states in solution.
No experimental structural comparison of HSPA8 SBD in the apo and peptide-bound state is available. However, there is strong NMR evidence that the Hsc70 SBD in the apo state is even more dynamically disordered than the DnaK SBD in the apo state (unpublished).
In summary, these less-than-perfect comparisons between different experimental structures reinforce each other, and it seems safe to conclude that the human Hsp70 domains are differently docked in ATP and ADP states, and that NBD subdomains rotate with respect to each other upon hydrolysis of the ATP gamma phosphate. Nature seems to use these varying surfaces to modulate the energetics of Hsp70 conformational equilibrium by co-chaperones, e.g.by DnaJ. Further, it seems likely that more interactions, which are not yet fully investigated or described, will be found to exploit these dramatic differences as well.
COMPLEXES WITH NEFS
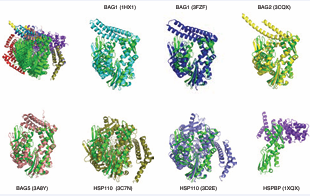
The next step in building the structural picture of how chaperone complexes function is to examine the interaction between Hsp70s and the NEFs. This interaction is emblematic of the protein-protein contacts that form the next “layer” of dynamics and allostery in the system and it has been extensively characterized by X-ray crystallography. The BAG proteins all bind to the “rear” of lobes IB and IIB and stabilize an open structure of the nucleotide binding cleft[16], providing a explanation for the nucleotide exchange mechanism (Figure 4).
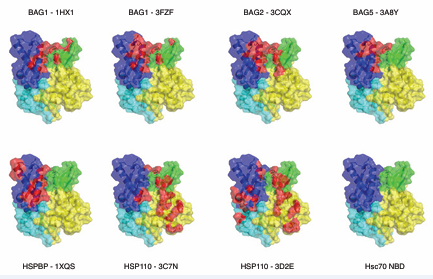
Figure 4: Comparison of the perturbation of Hsp70 NBD by nucleotide exchange factors. The HSPA8NBD in ADP state (3HSC) is shown in green. The structures are aligned on subdomain IIB. Top left: all structures compared. The other panels show the individual structures. Bag1 (1HX1), Cyan; Bag1 (3FZF), blue; Bag2 (3J8F), yellow; Bag 5 (3A8Y), salmon; Hsp110 (3C7N), slate; Hsp110 (3D2E), olive; HSBP (1XQS), purple.
The Hsp110 proteins engage a much larger surface of HSPA8, still including lobes IB and IIB[14,15]. Figure (4) also shows that the perturbation of the NBD cleft-opening varies significantly between the different NEFs. Whether such differences in extend of cleft opening also occur in solution is currently unknown. Similarly, the different BAG domains seem to have distinct orientations of their BAG helical bundles with respect to the HSPA8 NBD, but it isn’t yet clear if this reflects a meaningful divergence or if packing interactions are skewing the structures. The crystal structure for the HSPBP1 complex contains only NBD lobe II [17]. It can therefore not really explain the mechanism of nucleotide exchange, but it does indicate that, at least in lobe II, that the interaction surface is more extensive than that in the BAG series. No solution studies were published for the Hsp70 / NEF interactions. However, good quality NMR data could be collected on HSPA8 NBD in complex with BAG1, whereas addition of HSPBP1 to a solution of HSPA8NBD caused massive precipitation (unpublished results). This appears to indicate that HSPBP1 perturbs the NEF structure much more then than BAG1 does. Put together, the interaction surfaces of the different NEFs (i.e. HSPBP1, BAGs, Hsp110) on the Hsp70 proteins have some similarities, but are also different (Figure 5).
Figure 5: Interactions surfaces between Hsp70s and the different NEFS. The color coding is: Yellow: subdomain IA; Blue, IIA, Green, IB, Purple, IIB. In red are those NBD atoms (including H) that are within 3 Å of NEF atoms (including H).
In Figure (5), the orientation is 180 around the vertical axis as compared to Figure (4) to best illustrate the differences between the surfaces. Another way to look at the difference is by summarizing the residues that make contact in the structures (Table 1).
|
Table 1. Summary of HSPA8 NBD contacts with NEFSa |
|||||||
|
HSP110 (3C7N) contacts |
HSP110 (3D2E) contacts |
HSPB (1XQS) contacts |
BAG1 (1HX1) contacts |
BAG1 (3FZF) contacts |
BAG2 (3CQX) contacts |
BAG5 (3A8Y) contacts |
HIP (4J8F) contactsb |
|
H23 |
Q22 |
R247 |
T45 |
D46 |
Q33 |
A60 |
K25 |
|
K25 |
H23 |
K248 |
D46 |
N47 |
N35 |
L61 |
D32 |
|
E27 |
K25 |
H249 |
N57 |
A60 |
N57 |
K257 |
Q33 |
|
A30 |
D32 |
K250 |
A60 |
M61 |
Q58 |
R258 |
N35 |
|
D32 |
Q33 |
R258 |
M61 |
R258 |
A60 |
R261 |
N57 |
|
Q33 |
G34 |
R262 |
N62 |
R261 |
M61 |
R262 |
Q58 |
|
G34 |
R36 |
T265 |
R258 |
R262 |
R258 |
T265 |
L61 |
|
R36 |
L50 |
E268 |
R261 |
T265 |
R261 |
R269 |
Y134 |
|
L50 |
A54 |
R269 |
R262 |
R269 |
R262 |
I284 |
R258 |
|
A54 |
N57 |
A270 |
T265 |
S281 |
T265 |
D285 |
R262 |
|
N57 |
Q58 |
R272 |
R269 |
E283 |
A266 |
S286 |
R269 |
|
M61 |
L61 |
T273 |
S281 |
I284 |
R269 |
D292 |
T273 |
|
A133 |
A133 |
S277 |
I282 |
D285 |
E283 |
Y294 |
S281 |
|
Y134 |
Y134 |
Q279 |
E283 |
S286 |
I284 |
|
E283 |
|
R258 |
R262 |
A280 |
I284 |
G290 |
D285 |
|
D285 |
|
R269 |
T273 |
S281 |
D285 |
D292 |
S286 |
|
D292 |
|
R272 |
S276 |
L282 |
S286 |
Y294 |
D292 |
|
Y294 |
|
T273 |
T278 |
E283 |
G290 |
|
Y294 |
|
|
|
S276 |
Q279 |
D285 |
D292 |
|
|
|
|
|
T278 |
S281 |
D292 |
Y294 |
|
|
|
|
|
S281 |
D285 |
Y294 |
|
|
|
|
|
|
E283 |
S286 |
|
|
|
|
|
|
|
D285 |
T298 |
|
|
|
|
|
|
|
S286 |
R299 |
|
|
|
|
|
|
|
G290 |
A300 |
|
|
|
|
|
|
|
T298 |
R301 |
|
|
|
|
|
|
|
R299 |
E304 |
|
|
|
|
|
|
|
A300 |
K348 |
|
|
|
|
|
|
|
R301 |
D352 |
|
|
|
|
|
|
|
E303 |
|
|
|
|
|
|
|
|
E304 |
|
|
|
|
|
|
|
|
R342 |
|
|
|
|
|
|
|
|
aHsc70 residues within 3 Å of any residue of the co-chaperone, any atoms, including hydrogens. bCrystal contacts between HSPA1 NBD and HIP as described in Li et al. [44]. |
|||||||
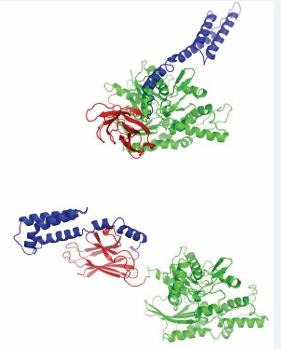
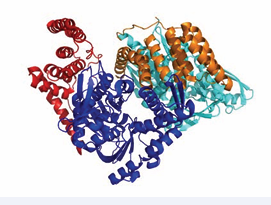
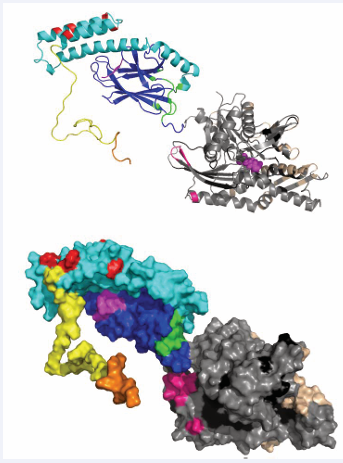
All surfaces are extensive, and will be quite difficult to inhibit. In addition, the surfaces overlap considerably so that it will be difficult to affect NEF – Hsp70 interaction with any specificity between NEFs. The possible exceptions are HSPA8 residues 45-47, that are involved in the BAG1 interaction only. As mentioned above, HIP is an “anti-exchange” factor, which stabilizes the Hsp70 ADP-state, via interactions between Hsp70 and the middle domain of HIP (residues 77-247) [43]. Although a full-length structure is not available, X- ray structures of the domains have provided some clues as to how it might work. For example, a fusion between HSPA1 NBD (1-383) and the HIP middle domain (77-247) without a linker was crystallized [44]. In that structure, the “cis” interface between HSPA1 and HIP is dictated by the short connection between the domains, and may not be relevant. However, a serendipitous “trans” crystal contact between HSPA1 of one fusion with HIP of another, resembles the interface of BAGs with HSPA8 [44] (Figure 6).
Figure 6: HSPA1 NBD (blue and cyan) linked to HIP middle domain (red and orange). The crystal contact between the blue NBD and orange HIP may constitute a natural complex.
This contact may explain the HIP-BAG antagonism. In addition, the NBD in this crystal structure is more closed than Hsc70 in the ADP state; hence, it seems that HIP closes the NBD cleft to stabilize the nucleotide-bound state (both ADP and ATP).
While the focus of this section has been on NEF interactions at the Hsp70 NBD, some of the NEFs seem to operate by multivalent contacts. For example, a crystal structure between DnaK NBD and the bacterial NEF, GrpE, suggested that NEFs could interact with the SBD as well [45]. In that structure, the long helical bundle of GrpE lies along the DnaK lobe I, and points in the direction of the SBD. Indeed, the Valpuesta and Mura groups have shown using biophysical methods that there is an interaction between a disordered region at the terminus of the helical region and the DnaK SBD [46]. In this model, GrpE directly competes with clients, helping to accelerate their removal. However, not all clients may work this way, as σ-32 has been shown to be released from DnaK GrpE in a way that is concerned with nucleotide release [47]. Very recently it was shown that the N-terminal domain of BAG 3, outside the BAG domain, interacts with the HSPA8 SBD [12]. The interaction is likely with the hydrophobic cleft, in a manner somewhat analogous to what was reported for DnaK [46]. Hence, some of the mammalian NEFs may also help substrate release in a bi-functional manner: stimulation of ATP binding and thereby increasing the substrate off-rate as well as direct competition with the substrate. Indeed, it was shown the isolated BAG domain from BAG-3 was unable to release a peptide client from HSPA1 on its own [12], suggesting that the multivalent contact is necessary. These observations contribute to a growing model in which chaperone interactions within the co-chaperone complexes are more nuanced than previously imagined, especially in the mammalian systems.
Complexes with J proteins
In contrast to the Hsp70 – NEF interaction, the interaction between Hsp70 and J-proteins has been much more elusive. The J-proteins are defined by the presence of a J-domain, which is a three-helical bundle of ~70 residues containing an invariant HPD loop between helices 2 and 3 [48]. Human HdJ1 contains a J-domain, a long linker, and one or more substrate-binding domains, and a dimerization domain [49]. Some J-proteins contain a J-domain connected to quite different protein-protein interaction domains, likely accounting for functional specificity [13]. The isolated J-domain has been shown to stimulate DnaK ATP hydrolysis, but does bind less tightly to the DnaK proteins than full-length J-proteins[50].Besides the conserved HPD loop, the J-domains display conserved lysines and arginines on helix 2.Mutagenesis of the HPD loop and / or the Lys/Arg results in loss of DnaJ stimulation of Hsp70 ATP hydrolysis, suggesting a possible involvement in the interaction or positioning of the proper residues. Mutagenesis of DnaK suggest that corresponding residues between lobes IA and IIA may be the opposing surface that makes contact with the HPD [51] however, this area on DnaK also mediates the NBD-SBD contacts in the ATP state, making the results difficult to interpret.
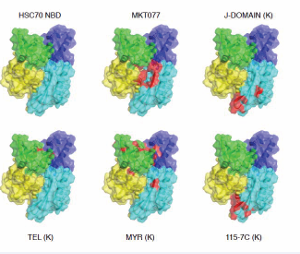
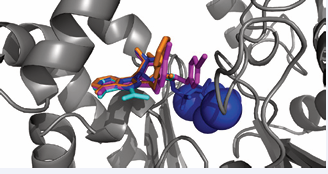
No crystal structure between an isolated J-domain and an Hsp70 has been reported, but a structure of a J-domain and the HSPA8 NBD covalently linked by a designed cross-link was published. That structure showed a proximity of residues in and around the designed cross link [52]. Mutation of some of these residues does disrupt J-protein driven allostery, but again these results are complicated by the roles of the lobe IA and lobe IIA region in many other aspects of the cycle. More recent solution NMR experiments may have revealed why it is has been so difficult to characterize the Hsp70 / J interactions: the affinity of the isolated J-domain (1-72) for wt-DnaK (ADP) is relatively weak (16 µM), while the binding surface on DnaK is found to be an extended area of negative electrostatic potential to which the positively charged helix II of DnaJ binds in a diffusive manner [53]. The center of this rather ill-defined area is at DnaK residue Asp 211/214 (DnaK/HSPA8), located at the periphery of IIA (see Figure 7 top-right).
Figure 7: Function-modifying interactions with the HSPA8NBD (3HSC). Subdomain IA: yellow; IB: green; IIA: cyan; IIB: blue. MKT077 interaction with HSPA8 from Rousaki et al. [71]. The following interactions were all obtained for the E.coli DnaK NBD, displayed on the Hsc70 NBD. DnaJ-J domain with DnaK from Ahmad [53]; Myricetin with DnaK from Chang et al. [68], 115-7c with DnaK from [67], Telmisartan with DnaK [69].
The DnaK and DnaJ contact areas correlate with those indicated by mutagenesis, except for the HPD loop. No evidence was found for the proximity of this loop to DnaK in the study by Ahmad [53], although transient contacts could not be excluded. DnaJ’s helix II also delocalize to a negative area on the “face” of the SBD [53]. DnaJ stimulates Hsp70 ATP hydrolysis, but binds more strongly to DnaK in the ATP state than to the ADP state [50]. Hence, there is no free-energy potential that would drive DnaK in hydrolysis transition state to the ADP state. Perhaps the (modest) ATP hydrolysis stimulation is caused by remote electrostatics. The hydrolysis product ADP.HPO4 with a total 5- charge is more negatively charged than ATP (4-). Hence, the presence of a positively charged DnaJ could stabilize the products of hydrolysis.
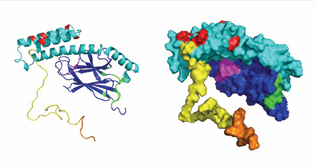
TPR-protein interactions: Hsp70 proteins utilize the C-terminal sequence of IEEVD to interact (Figure 8)
Figure 8: HSPA8 model based on DnaK SBD in the ADP state (1DKX), extended with an unstructured tail comprising residues 605-646. NBD, grey; SBD BETA domain: Blue; SBD LID domain: cyan; tail: yellow. In magenta, substrate peptide NRLLLTG and nucleotide ADP. Interaction sites: all NEFs: beige; all non-ATP site Hsc70 and E. coli DnaK NBD binding compounds with activity: black. CHIP: orange from [57]; Phosphoserine lipids: red [62]; J-domain – E. coli DnaK interaction from [53] pink; E. coli DnaK interaction with PET-16 (4R5G): green.
with the TPR proteins HOP, CHIP, PP5, DnaJC7, FKBP51, and FKBP52 [54].Mutagenesis in the IEEVD sequence shows loss of binding and alanine scanning has shown the residues important in the interaction. Further, details of the interaction of the CHIPTPR domain with GTIEEVD [55]and PTIEEVD [56] peptides were elucidated by X-ray crystallography (See Figure 9).
Figure 9: The interaction of the CHIP TPR domain with the HSPA8IEEVD C-terminal peptide (3Q49). CHIP is as a cartoon while IEEVD is as sticks. Color coding: Grey, polar; Green; hydrophobic; Red, negative; Blue positive.
The interaction is clearly stabilized by electrostatics involving HSPA8’s E643, D646 and the COO- terminus. But there is more to it: HSPA8’s I641 is tightly sandwiched between CHIP residues F98 and F131, andHSPA8V645 contacts CHIP’s Phe 37 and Leu 68. Rather unexpectedly, HSPA8’s E644 is pointing outside of the TPR pocket. NMR studies by Zhang et al. [56], indicate that CHIP interacts besides the C-terminal tail also with the HSPA8SBD itself. NMR studies of the interaction of full-length CHIP with full length HSPA8 [57], on the other hand, show no evidence of the latter interaction, although transient touches cannot be excluded. The hallmark of the interaction as determined by the latter NMR studies, show CHIP to be dynamically tethered to Hsc70 through the IEEVD clamp, with HSPA8 residues 610-638 remaining flexible and unaffected by the presence of CHIP.
This type of scenario may be biologically advantageous [57], because the E2 ubiquitin ligase bound to CHIP can reach more residues of the HSPA8 substrate for ubiquitination than if the complex was rigid. The same group also demonstrated a dynamic tethered binding mode for the Protein Phosphatase 5, which also uses its TPR domain to interact with the HSPA8IEEVD sequence [58]. This seems equally desirable: a tethered phosphatase can dephosphorylate many more substrate residues than a statically bound one.
Complexes for autophagy
Lysosomes take up and degrade intracellular proteins in response to serum deprivation or other nutrient signalsin a process called autophagy. There are many types of autophagy, including micro-autophagy and macro-autophagy. By injecting a variety of proteins into fibroblasts, Dice [59] discovered that certain proteins containing a peptide sequences biochemically related to KFERQ are preferentially targeted for hydrolysis by a discrete mechanism. In subsequent work, his group determined that these KFERQ proteins were recognized by HSPA8 [31]. Further, it was discovered that the HSPA8-bound KFERQ proteins are(1) targeted to the LAMP2A receptor on lysozomes for internalization and hydrolysis, in a process termed Chaperone-Mediated Autophagy (CMA) [60] or (2) to late endosomes for complete or partial degradation called Endosomal microautophagy [61]. Protein cargo selection is mediated by the chaperone HSPA8 and requires the SBD of HSPA8 for electrostatic interactions with the endosomal membrane [61]. The intermolecular interactions needed for both processes are just recently being investigated.
It is clear that the KFERQ sequence does not resemble the prototypicalHSPA8substrate such as NRLLLTG, which is hydrophobic carrying a positive charge on the Nterminus. In ongoing NMR work, we observe spectacular changes in the NMR spectrum of the isolated HSPA8NBD upon binding of known substrates such as NRLLLTG, but we donot find any change upon adding KFERQ (unpublished results). Additionally, RnaseA, which is the prototypical CMA client[59], also fails to induce the strong perturbations as seen for NRLLLTG. Accordingly, as of now, the mechanism of recognition between the KFERQ sequence and HSPA8 remains elusive. However it is absolutely clear that this interaction is not the same as a canonical hydrophobic peptide SBD binding.
The endosomal membrane contains a large fraction of phosphoserine lipids, which carry a net negative charge [61] had previously determined that the HSPA8SBD is involved in the interaction between membrane and protein necessary for the ESCORT – mediated translocation. Very recently [62], using NMR and mutagenesis, narrowed the interaction site down to a cluster of positive charges on the HSPA8LID domain. This interaction site, quite different from any other known site, is shown in Figure 8. The NMR data show that HSPA8 remains mobile when bound to a large vesicle containing 100% DOPS. This indicates that the HSPA8-membrane interaction is diffusive, and likely very difficult to perturb with small compounds.
Finally, CMA involves interaction of a HSPA8-KFERQ substrate complex with the LAMP2A receptor located in the lysosomal membrane surface [60]. LAMP2A is integral membrane protein that serves as pore for cargo transport. The HSPA8-KFERQ substrate complex interacts with theLAMP2A C-terminal cytosolic peptide GLKRHHTGYEQ [60]. Recent NMR data demonstrates that the cytosolic tail of LAMP2A may bind HSPA8 and its cargo simultaneously [63]. However, it remains unknown how HSPA8 recognizes the LAMP2A receptor and it remains unclear how these interactions lead to CMA.
COMPOUND INTERACTIONS
One of the reasons for better understanding the structure and function of the Hsp70 complexes is to enable the discovery of chemical inhibitors. However, this class of chaperones is a significant challenge. Inhibiting the nucleotide binding with a competitive inhibitor is difficult because of the high conservation between the ATP-binding fold in the Hsp70 NBD and the fold in other abundant proteins, such as actin. Further, the tight affinity of Hsp70 for ATP and the high cellular concentration of ATP/ ADP make it difficult to effectively compete for this interaction. On the other end of the chaperone, any molecule capable of directly competing with the client-binding cleft of the SBD would, likewise, need to contend with the high concentration of possible clients. Moreover, such a molecule would need to be quite hydrophobic. On top of these problems, it is not clear how one might specifically target a single paralog of Hsp70, owing to their high homology.
Despite these obstacles, several groups have made progress in developing inhibitors. Here, we review these compound protein interactions, in much the same way as we reviewed the co-chaperone interactions. Again, the goal is to elucidate the broader concepts across different observations and groups.
A group at Vernalis, Inc. were the first to report nucleotide analogs that inhibit HSPA1 [64,65]. These molecules were developed through a fragment-based and structure-guided program, resulting in molecules that make contacts with key residues in the NBD (Figure 10).
Figure 10: The HSPA8NBD (3HSC) as a cartoon is in grey. ADP+Mg+Na+PO4 bound to the HSPA8 NBDare in blue; Compound SGV in cyan (5AQZ); compound GB8 in red (5AR0); compound 3FD (VER-155008) in magenta (4I08).
As designed, the nucleosides of the three analogues occupy the same location as ADP in HSPA8. Only VER-155008 contacts other parts of the protein and offers some hope for being Hsp70 chaperone selective. Its properties were discussed in detail by Schlecht [66], who showed it to be a competitive ATP binding inhibitor.
Hsp70 has evolved to use a great many of its surfaces and crevices to engage in both intra- and inter-molecular interactions. This “economical” approach gives hope that one can affect the protein using molecules that bind to less obvious, allosteric regions. One of the first compounds identified to inhibit DnaK’s ATP hydrolysis was 115-7C, was found to locate to the interface of DnaK that is involved in J protein interactions[67] (Figure 7). Clearly, this molecule modulates Hsp70 function in an indirect way by interfering with J-domain binding (Figure 7). This binding site may allow the compound class to have eventually access better selectivity, because of the relatively lower conservation in that region. While 115-7c acts on the J-protein interface, several other Hsp70 modulating compounds are likely to be involved in modulating Hsp70’s NEF interactions [68,69] (Figure 7). A third class of allosteric inhibitors is based on the rhodocyanine, MKT077. MKT077 had been in Phase I clinical trial in the 1990s as a very promising, high-therapeutic window, tumor suppressor but the trial failed because of the compound’s renal toxicity [70]. By NMR, the binding site of MKT077 was found to be a deep pocket on the HSPA8 NBD between domains IA and IIA (Figure 7), allocation that is adjacent to the nucleotide-binding site [71]. Significantly, the binding occurs only in the ADP state, which could be rationalized because this pocket is occluded in the ATP state of the protein [42]. Hence, serendipitously, an allosteric modulator was found that drives HSPA8 to the ADP state / conformation where it has high affinity for its substrates. Thus, MKT077 can stall the HSPA8 refolding cycle in the substrate-bound state, increasing the chance that the complex will be recognized by CHIP and guided to the proteasome. This mechanism may be underlying the now well-documented ability of MKT077 derivatives to help clear tangles of aberrant tau protein in Alzheimer’s model cells [72].
Recently, fragment-based screening monitored crystallography was used to find additional binding sites in proteins[73]. Amidst 20 other proteins, HSPA2 NBD was also included in the test, and 4 compounds were obtained that bind to HSPA2 in three different binding locations (Figure 11).
Figure 11: Non-canonical fragment binding sites on HSPA2 NBD, not tested for any activity [73]. Cyan, compound PZA (5FPD.pdb); Red, compound IWT (5FPM.pdb); Magenta: compound KYD (5FPN.pdb).
Compound IWT (red) is at an interesting location that could modulate the NBD-SBD allostery. The other two locations donot change conformation between ATP and ADP state, are not involved in chaperone-co-chaperone interactions, and thus seem less interesting.
At present, two non-peptide compounds were identified that interact with the Hsp70 SBD. The inhibitor 2-phenylethynesulfonamide (PES) interacts with the SBD of HSPA1 [74]. Schlecht [66] tested the compound in folding and binding assays, and found that its activity was detergent-like. Another compound, PET-16, affects Hsp70 function by allostery [75]. The hydrophobic compound nestles itself within the hydrophobic core of the SBD, at a location that likely disturbs the allosteric communication between the substrate in the SBD and NBD interface on the SBD (Figure 12).
Figure 12: Summary of all interaction sites discussed on a homology model of full – length HSPA8 in the ADP/peptide-bound state, based on DnaK in the same state (2KHO). NBD, grey; SBD beta domain, blue; SBD LID, cyan; tail, yellow. ADP and substrate are in magenta. Combined NEF sites in beige; J site in pink, CHIP site in orange; phosphoserine lipid site in red; PET-15 site in green; the active compound sites in black.
This binding site was already discovered by Wang [76] as the only free peptide binding site in a SBD construct that was self-associated in its primary binding site.
One of the fascinating aspects of the small molecule interactions with Hsp70 is the diversity of binding sites. The Hsp70 residues that interact with the different chemicals discussed are listed in Table (2),
|
Table 2. Summary of HSPA8 contacts with CHIP, J and various chemicals |
||||||||||||
|
CHIPa
|
MKT-077b |
DOPSc |
J-domaind |
PET-16e |
Myricytinf |
115-7cg |
Telmisartanh |
|||||
|
T641 |
R76 |
K569 |
D208 |
E213 |
L392 |
L394 |
K55 |
K56 |
T173 |
T177 |
R56 |
N57 |
|
I642 |
A148 |
K573 |
D211 |
D214 |
D393 |
D395 |
R56 |
N57 |
A174 |
A178 |
M89 |
W90 |
|
E643 |
Y149 |
K583 |
G212 |
G215 |
V394 |
V396 |
Q57 |
Q58 |
A176 |
A180 |
F91 |
F92 |
|
E644 |
F150 |
K589 |
T215 |
I216 |
T395 |
T397 |
A58 |
V59 |
L177 |
I181 |
R261 |
R264 |
|
V645 |
T222 |
K601 |
F216 |
F217 |
P396 |
P398 |
V59 |
A60 |
G180 |
G184 |
|
|
|
D646 |
A223 |
|
E217 |
E218 |
L397 |
L399 |
L66 |
V67 |
L181 |
L185 |
|
|
|
|
G224 |
|
D326 |
D323 |
L399 |
L401 |
F67 |
F68 |
D182 |
D186 |
|
|
|
|
D225 |
|
|
|
I418 |
I420 |
G74 |
G75 |
K183 |
K187 |
|
|
|
|
T226 |
|
|
|
Q442 |
E444 |
I88 |
H89 |
N187 |
E192 |
|
|
|
|
H227 |
|
|
|
I478 |
I480 |
M89 |
W90 |
R188 |
R193 |
|
|
|
|
L228 |
|
|
|
A480 |
A482 |
F91 |
F92 |
T189 |
N194 |
|
|
|
|
|
|
|
|
D481 |
N483 |
K92 |
M93 |
A191 |
L196 |
|
|
|
|
|
|
|
|
G482 |
G484 |
V103 |
V105 |
V192 |
I197 |
|
|
|
|
|
|
|
|
A503 |
N505 |
K106 |
K108 |
S203 |
S208 |
|
|
|
|
|
|
|
|
S504 |
D506 |
A111 |
S113 |
I204 |
I209 |
|
|
|
|
|
|
|
|
|
|
N222 |
A223 |
I205 |
L210 |
|
|
|
|
|
|
|
|
|
|
G223 |
G224 |
D211 |
D214 |
|
|
|
|
|
|
|
|
|
|
E230 |
E231 |
G212 |
G215 |
|
|
|
|
|
|
|
|
|
|
I237 |
V238 |
T215 |
I216 |
|
|
|
|
|
|
|
|
|
|
M259 |
V260 |
F216 |
F217 |
|
|
|
|
|
|
|
|
|
|
Q260 |
R261 |
E217 |
E218 |
|
|
|
|
|
|
|
|
|
|
L262 |
L263 |
|
|
|
|
|
|
|
|
|
|
|
|
E267 |
E268 |
|
|
|
|
|
aFrom NMR intensity perturbations of human wt-HSPA8 bFrom NMR Chemical Shift Perturbation (CSP) in human HSPA8(1-386) cFrom NMR CSP in human HSPA8(387-604) and mutagenesis of full-lengthsHSPA8 dFrom Paramagnetic Relaxation Enhancement NMR for E.coli DnaK(1-608). Left column: DnaK count, right column: human HSPA8 count. e3A proximities between compound and E.coli DnaK in the crystal structure (4R5G). Left DnaK count, right HSPA8 count. fFrom NMR CSP in E.coli DnaK(1-388) Left DnaK count, Right: HSPA8 count gFrom NMR CSP in E.coli DnaK(1-388) Left DnaK count, Right: HSPA8 count hFrom NMR CSP in E.coli DnaK(1-388) Left DnaK count, Right: HSPA8 count |
||||||||||||
illustrating this point. Much like the co-chaperone interactions that are spread across the entire surface of the protein, the compound interaction sites seem to be similarly distributed. The “best” ones and their effects on chaperone functions in disease are only beginning to be made clear.
CONCLUSIONS
In conclusion, the Hsp70s have evolved to utilize many different interfaces to accommodate many different binding partners. Figure (8) serves as an overview of the different binding sites discussed here. Even the chaperone itself uses completely different interfaces between its domains (NBD, SBD, LID) in the different allosteric states. While it is likely that most of the intermolecular interfaces have now been identified, there are still several known unknowns that need to be hunted down: (1) the interaction of NEFs with SBD (2) the true interaction of HIP (3) possible Hsp70-Hsp70 interactions when bound to substrate (4) the interaction with DnaJ in the ATP state (5) the interaction of Hsp70 proteins with autophagy (KFERQ proteins) clients (6) the interaction with the LAMP2A receptor. Likely there are several more for now unknown interfaces with for now unknown Hsp70 partners to be chased in the future.
ACKNOWLEDGEMENT
This work was supported by NIH grant 5-R01 NS-059690-01-08.