Understanding the Role of Point Mutations in Rbfa of Escherichia coli: Insights from In silico Modeling and Docking Studies
- 1. Department of Biological Sciences and Bioengineering, Indian Institute of Technology Kanpur, India
Abstract
Ribosomal binding factor A (RbfA) and YjeQ, ribosome small subunit-dependent GTPase A (RsgA) are two important ribosome assembly factors among many factors involved in maturation of 30S ribosomal subunit. They exhibit a functional interplay, both at the genetic and molecular levels during the maturation of 30S ribosomal subunit. Recently, an abnormal ribosome profile with low ratios of 70S ribosomes to the 50S and 30S subunits was observed in ΔYjeQ cells as compared to wild type. This results in the death of ΔYjeQ cells due to improper ribosomal maturation. In order to overcome this effect, the bound RbfA should be released from 30S subunit complex upon ΔYjeQ binding. Surprisingly, the cells that are rescued from death are known to have point mutations at six positions in RbfA. These mutant versions of RbfA were speculated to show poor binding to ribosome. In the present study, we explain the possible role of the mutations in RbfA through in silico modeling and docking. Our results indicate that the mutations play a major role in gain of new function by the RbfA compared to the wild type RbfA protein. These findings further suggest that the point mutations weaken the efficiency of interaction of RbfA with 30S subunit thereby aiding in its release after the completion of the function.
Keywords
RbfA; YjeQ; Ribosome; Ribosome biogenesis; Point mutation
CITATION
Gunasekhar B, Malik SC (2018) Understanding the Role of Point Mutations in Rbfa of Escherichia coli: Insights from In silico Modeling and Docking Studies. JSM Cell Dev Biol 6(1): 1025.
ABBREVIATIONS
Rbfa: Ribosomal Binding Factor A; Gtpase: Guanosine Triphosphate Hydrolase; Rsga: Ribosome Small Subunit Dependent Gtpase A (Yjeq); Rimm: Ribosome Maturation Protein; MSA: Multiple Sequence Alignment; R: Arginine; H: Histidine; P: Proline; L: Lysine; G: Glycine; D: Aspartic Acid; E: Glutamic Acid; RNA: Ribonucleic Acid; Rrna: Ribosomal RNA.
INTRODUCTION
The ribosome biogenesis can happen in cytoplasm [1] as well as nucleolus [2]. The former takes place in prokaryotes [1], while the later in eukaryotes [2-4], although eukaryotes can have cytoplasmic ribosome biogenesis. It is the process of making ribosomes which is necessary for cell growth and proliferation [5], where they play a key role in translation. During ribosome biogenesis several assembly factors and binding proteins associate to regulate the process [4]. At the same time, release of assembly factors is vital for the maturation of ribosomal subunits [3].
Binding of assembly factors to ribosomal subunit is necessary and crucial during maturation [6]. Some assembly factors are phenotypically similar, while some are functionally similar. In bacteria, RsgA, RbfA, Era and RimM are a group of phenotypically similar assembly factors involved in 30S subunit maturation [7,8].The RsgA belongs to a class of GTPases as it exhibits GTPase activity in order to overcome the energy barrier during maturation [1,9-11]. It is also referred as YjeQ [11,12]. Its N-terminal region contains the oligosaccharide binding fold (OB fold) and the zinc binding takes place at the C-terminal end [13 15]. This activity of RsgA is subunit dependent [12]. The ratio of 70S ribosome to its subunits is altered when RsgA is absent or mutated. In some cases, its absence can be overcome by other assembly factors.
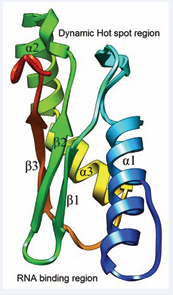
Further, RsgA was shown to be crucial as it aids in releasing RbfA from the 30S subunit. This step is vital for late stage maturation of 30S ribosomal subunit. RbfA is a protein of functional importance which is necessary for efficient processing of 16S rRNA [16,17]. It consists of mainly K homology type II (KH type II) domain, responsible for binding to nucleic acids. The domain constitution is α1-β1-β2-α2-α3-β3 (α+β fold) fold topology (Figure 1). The fold topology is arranged such that it has a strong electronegative field at one end and a strong electropositive field at the opposite end (Figure 1) [16].
Figure 1: Structure of RbfA protein (PDB ID: 1KKG) showing dynamic hot spot region and positively charged region.
The bipolar nature of RbfA could help in binding and release from the ribosomal subunit as opposite electrostatic fields on either side can interact with YjeQ. Interestingly, it can only bind to 30S subunit. It can neither bind to the other subunit, nor to the whole 70S ribosome [16]. The wild type RbfA strongly interacts with 30S subunit and hence it needs YjeQ to release it after completing the function [10]. Null mutants of these two assembly factors are similar, structurally [17] as well as functionally [16].
Recently, an abnormal ribosome profile with low ratios of 70S ribosomes to the 50S and 30S subunits was observed in ΔYjeQ cells compared to wild type [10,16]. This resulted in the death of ΔYjeQ cells due to improper ribosomal maturation. This is due to the inability of the ΔYjeQ to release the bound RbfA from 30S subunit complex upon its binding [10]. Surprisingly, some cells were found to overcome this effect and are rescued from death. Interestingly, those cells were found to contain point mutations at six different positions in RbfA which result in poor binding of RbfA to ribosome [10].
In this study, we intend to understand how these point mutations nullify the effect shown by ΔYjeQ. For this, five mutants of RbfA were modeled and the mutant RbfA structures thus obtained were docked on to the 30S subunit to understand the efficiency of binding of mutant versions in comparison with the native RbfA. These findings suggest that the point mutations weaken the efficiency of interaction of RbfA with 30S subunit. This might be the reason for easy release of RbfA in ΔYjeQ cells.
METHODOLOGY
Multiple Sequence Alignment (MSA)
MSA was carried for full length RbfA protein sequences from 14 RbfA family members. For this, we used the blastp tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) that compares protein queries with protein databases [18]. Using this, the respective FASTA sequences were collected for all the 14 RbfA family members. These sequences were subjected to MSA using muscle server (http://www.ebi.ac.uk/Tools/msa/muscle) with all the parameters set to default values. We have also used Jalview, a freely available MSA tool for analyzing the data.
Modeling of mutant RbfA
Point mutations were induced at respective positions i.e.R10H, P30L, G77D, G84E and D100G in RbfA protein [10]. This was achieved by replacing an amino acid in wild-type RbfA primary sequence at respective position with an amino acid observed in case of mutant forms. The primary sequences with point mutations that were obtained as a consequence were then subjected to modeling by using SWISS-MODEL, which is a fully automated and freely available protein structure homology modeling server (http://swissmodel.expasy.org) [19-21]. For this, the wild type RbfA (PDB ID: 1KKG) was used as a reference [22]. The resultant structures were analyzed for changes in secondary structure, intra and/or inter-molecular interactions and related data using UCSF chimera (http://www.cgl.ucsf.edu/ chimera) program [23-27].
Docking mutant RbfA on to 30S subunit
The mutant versions of RbfA were docked on to the 30S ribosomal subunit by using Hex Server protein docking and molecular superposition program (http://hex.loria.fr/hex.php) [28]. In order to maintain uniformity, default parameters were used in all the cases. The results were analyzed for any changes in the interactions of mutant RbfA proteins with 30S ribosomal subunit in comparison with that of the wild type RbfA protein. This was also analyzed by using UCSF Chimera (http://www.cgl. ucsf.edu/chimera) software [23-27].
RESULTS AND DISCUSSION
The process of ribosomal biogenesis involves coordination of many subcellular events including transcription of genes that encode for ribosomal RNAs (rRNAs), associated enzymes and structural proteins, modification of rRNAs, post translational modification of the proteins and the assembly of these proteins with the rRNAs. Although many factors are involved in the ribosome biogenesis, no factor except RNases and modifying enzymes has been reported with definite functions [1]. RbfA, YjeQ (RsgA), Era and RimM are some of the factors involved in 30S subunit assembly [7,8]. Interestingly, RbfA and YjeQ are found to exhibit functional interplay during the 30S subunit maturation at the genetic as well as molecular level [10].
The strongly negative end of RbfA contains a “dynamic hot spot” region (Figure 1) in the vicinity of a highly conserved Ser39 residue that forms the β-bulge [29].The amino acid residues that are involved in the constitution of hot spot region include Gln24 Arg25, Lys28-Asp29, Thr37-Gly40, Val54-Thr55, Thr57 and Tyr99-Asp100. The opposite end is strongly positive because of the presence of basic residues – Arg80, Lys85, Arg88, Arg90, Arg7, Arg10 and Arg45 which are likely to be involved in interactions with rRNA (Figure 1) [29]. Apart from these regions, RbfA also contains many regions with high internal conformational dynamics and flexibility [29].
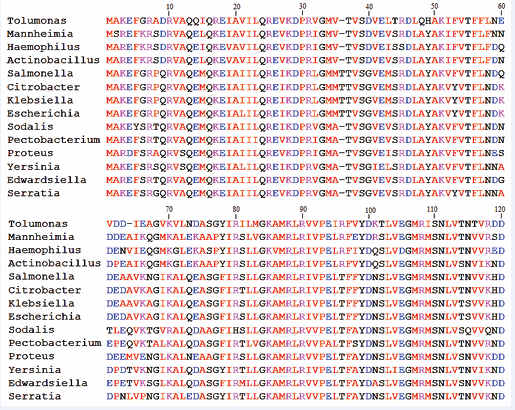
Therefore, multiple sequence alignment (MSA) analysis was carried out for this protein (Figure 2) to understand the significance of such regions.
Figure 2: Multiple sequence alignment data showing high sequence conservation among fourteen RbfA family members. First hundred and twenty amino acid residues were shown here due to some variations in total length of the protein across different organisms.
The results of MSA showed highly conserved signature sequences across the observed organisms of RbfA family. This indicates a strong biological importance of this protein. High sequence conservation is observed at many positions mostly with respect to the amino acids present in and around the dynamic hot spot region and the region of RbfA that binds to RNA or 30S ribosomal subunit (Figure 2). These findings suggest that there exists an evolutionary pressure in maintaining the sequence and structure of this region [29]. Hence, any alteration or mutation in the primary sequence may completely change the properties of RbfA.
Binding of RbfA to the 30S subunit helps in binding of YjeQ. This association of YjeQ promotes the release of RbfA from the ribosomal subunit thereby aiding in 30S subunit maturation. In E. coli, the null mutants for either RbfA or YjeQ were found to exhibit similar phenotype such as - accumulation of 17S Rrna [17], an abnormal ribosome profile with low ratios of 70S ribosomes to the 50S and 30S subunits individually as well as a reduction in cellular growth rate [16]. This might be the reason for cell death that was observed in ΔYjeQ cells as compared to wild type. This effect is due to the inability of the ΔYjeQ to release the bound RbfA from 30S subunit complex [10].
Surprisingly, some cells were found to overcome this effect and are rescued from death. These cells contain point mutations at six different positions in RbfA [10]. These mutations might lead to poor binding of RbfA to 30S subunit thereby aiding in its release either directly or indirectly. Hence, we intended to test this hypothesis based on in silico modeling and docking studies. For this, we used the template based homology modeling tool, SWISS-MODEL (http://swissmodel.expasy.org) [19-21,30]. It is a fully automated online tool that uses suitable templates for the prediction of more accurate models of protein structures [21]. Further, the accuracy of the generated models was independently evaluated by the CAMEO project (http://cameo3d.org) [31]. Also, the structural prediction was made for the sequences which are point mutants of RbfA, a 133 residue protein. Hence, it is believed that the structures predicted by SWISS-MODEL should be accurate and might serve the purpose of our study.
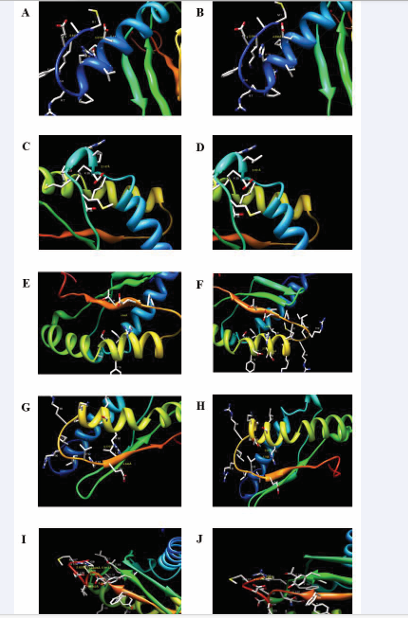
The modeled mutant structures were analyzed using UCSF chimera (http://www.cgl.ucsf.edu/chimera). Some significant changes in the local structure of the protein around the site of mutation were observed (Figure 3A-J).
Figure 3: Structures of wild-type (left column) and modeled mutants (right column) of RbfA. [A, B], [C, D], [E, F], [G, H] and [I, J] represent the mutations R10H, P30L, G77D, G84E and D100G respectively. These images show the altered interactions or local structural changes due to respective mutations.
This was found to be true in case of R10H mutation where histidine residue showed stronger interactions with the side chain of glutamic acid residues present at the fourth and fourteenth position when compared to arginine residue (Figure 3A& 3B). As a result there might be a small contraction within the helix at the N-terminal region as well as between the N-terminal loop and helix. Apart from this, the positive end of RbfA, where this mutation is present, is involved in interaction with rRNA. Generally, the side chain of arginine residues are involved in interaction with negatively charged non-protein atoms, possibly resulting in better binding of RbfA to 30S subunit [32,33].This is due to the presence of an ideal and positively charged guanidinium group that can be readily protonated on its side chain [33]. Also, its ability to form multiple hydrogen bonds makes it suitable for binding negatively charged phosphate groups [33].Thus the R10H mutation enhanced the intra-molecular interaction thereby compromising the interaction of RbfA with rRNA. This compromised inter molecular interactions thus may aid in easy release of RbfA in presence of ΔYjeQ [33].
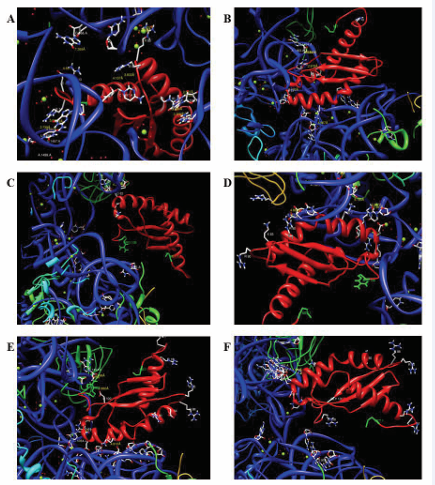
The R10H mutant was then docked on to the 30S subunit to analyze the efficiency of interaction between these two molecules. This was performed using Hex docking server. The protein docking on Hex Server is based on First Fourier Transform (FFT) powered by graphics processors. Simultaneously it uses two graphics processors making a 6D docking possible in just ~15s [28]. To our surprise, instead of the positive end as in the case of wild-type (Figure 4A), the negative end of RbfA, where the dynamic hot spot region is present, was found to interact with the 30S subunit (Figure 4B). The penetration of dynamic hot spot region resulted in weaker interactions between Arg108-G1494 (4.441Ao), Asp100-Arg108 (2.878Ao) Lys66-G966 (4.593Aoand 4.785Ao) and Asp61-C1397 (3.402Ao). This might be due to the repulsion between the negatively charged end of mutant (R10H) RbfA and the negatively charged rRNA present in the ribosome. Taken together, these studies suggest that the R10H mutation aids in the release of RbfA upon ΔYjeQ binding.
The R10H mutant was then docked on to the 30S subunit to analyze the efficiency of interaction between these two molecules. This was performed using Hex docking server. The protein docking on Hex Server is based on First Fourier Transform (FFT) powered by graphics processors. Simultaneously it uses two graphics processors making a 6D docking possible in just ~15s [28]. To our surprise, instead of the positive end as in the case of wild-type (Figure 4A), the negative end of RbfA, where the dynamic hot spot region is present, was found to interact with the 30S subunit (Figure 4B). The penetration of dynamic hot spot region resulted in weaker interactions between Arg108-G1494 (4.441Ao), Asp100-Arg108 (2.878Ao) Lys66-G966 (4.593Aoand 4.785Ao) and Asp61-C1397 (3.402Ao). This might be due to the repulsion between the negatively charged end of mutant (R10H) RbfA and the negatively charged rRNA present in the ribosome. Taken together, these studies suggest that the R10H mutation aids in the release of RbfA upon ΔYjeQ binding.
Figure 4: Docking images of wild-type and mutant forms of RbfA. Shown here are the interactions of wild-type (A), R10H (B), P30L (C), G77D (D), G84E (E) and D100G (F) forms of RbfA with the 30S subunit of ribosome.
In case of G77D (Figure 3F) and G84E (Figure 3H) point mutations, the net charge around the respective positions may be slightly changed towards negative. This effect should be prevalent in the latter case as the entire loop is rich in positively charged residues (Figure 3H). These mutations may lead to an abnormal interaction with nearby residues and may affect the binding efficiency of RbfA. This is more evident in case of G84E mutation as the positive residues in this region are involved in interaction with 30S subunit [29]. Upon docking, marginal interactions observed are Arg80-U956 (4.866Ao), Lys66-U965 (3.383 and 2.917Ao) and Lys70-U952 (5.185Ao) and Arg108-A1493 (3.043Ao), Arg108-1494 (3.994Ao), Lys70-G953 (4.812Ao) and Asp61-C1054 (1.337Ao) for G77D (Figure 4D) and G84E (Figure 4E) mutants respectively. These results together indicate that the binding is not as strong as the wild type (Figure 4A) and hence the mutant RbfA may easily get released from the ribosomal subunit.
Aspartic acid (D100), a strong negatively charged amino acid which constitutes the hot spot region may play an important role in maintaining the dynamic nature of the wild type RbfA (Figure 3I). Thus, when such charged amino acid is mutated (D100G) to Glycine (Figure 3J) which is uncharged, the dynamic nature of that region may get altered, in-turn changing the property of mutant RbfA. The docking results of D100G mutant (Figure 4F) showed similar interaction as that of P30L mutant. No proper interactions were observed except for a very weak interaction between Lys66 and A1492. These results clearly indicate that the interaction of D100G mutant RbfA with 30S subunit is quiet weak and hence may facilitate easy release of it from the ribosomal subunit. Thus the mutation at the dynamic hot spot region may change the entire property of binding.
CONCLUSION
From the above results, it is sensible that the positive region is involved in making strong interaction with the 30S subunit in the wild type RbfA. In native state, the highly positive end penetrates into the binding site of 30S subunit. Among many interacting residues, strong interactions were observed for Arg7 with G1401 and U1498, Arg10 with G966, Arg80 with G530 and C518, Arg88 with G963 and Arg90 with C1397 (Figure 4A). Hence, an additional factor may be required to release the native RbfA from 30S subunit. Also, the hotspot region is highly dynamic in nature. Therefore, any modification or mutation in these two regions is highly sensitive and may completely change the binding property of RbfA. Also, the mutations were found in the highly conserved regions of RbfA (Figure 2). This study clearly suggests that any modification in the highly conserved regions, the dynamic region and the positive region can completely change the binding property of RbfA. Finally, it seems reasonable to conclude that the point mutations induced gain-of-function by the mutated versions of RbfA. The mutations thus aid in the easy release of RbfA from the 30S subunit thereby promoting the 30S subunit maturation upon ΔYjeQ binding.
ACKNOWLEDGEMENTS
The authors would like to acknowledge all the freely available online bioinformatics tools used in the present study. Special thanks to Rituraj Niranjan for the guidance, motivation and valuable inputs for preparing the manuscript.