Proteomic Analysis of Diabetic Foot Skin Highlights Neutrophil Dysfunction and Identifies Potential Therapeutic Targets MDL 28170 and Amlexanox
- 1. Department of Endocrinology, Southeast University, China
Abstract
Diabetic foot (DF) and its complications represent a growing public health challenge, significantly impacting morbidity and mortality in the global diabetic population. The pathogenesis of DF is complex, involving multiple systemic and local factors that impede wound healing and exacerbate tissue damage. In this study, we conducted an in-depth proteomic analysis of diabetic foot skin (DFS) and non-diabetic foot skin (NFS) samples, revealing substantial differences in protein expression profiles. Principal component analysis (PCA) highlighted these differences, with a significant number of proteins differentially expressed, particularly those involved in neutrophil degranulation, coagulation, and angiogenesis. Further analysis identified 449 proteins with significant expression changes, 244 of which were upregulated in DFS. Functional enrichment using Metascape indicated a pronounced involvement of neutrophil extracellular traps (NETs) and related immune responses, which have been implicated in diabetic complications. To corroborate these findings, we integrated public gene expression datasets, identifying 18 key proteins linked to DF pathology. Molecular docking studies further identified MDL-28170 and amlexanox as potential therapeutic agents targeting these proteins, with strong binding affinities suggesting promising therapeutic efficacy. Our findings emphasize the critical role of neutrophil dysfunction in DF and offer new avenues for targeted treatment strategies. However, the study’s limitations, including a small sample size and the need for further in vivo validation, highlight the importance of ongoing research to translate these findings into clinical applications.
Keywords
Diabetic Foot; Proteomics; Neutrophil Extracellular Traps: Mdl-28170; Amlexanox; Therapeutic Targets; Wound Healing; Immune Dysregulation.
Citation
Yin H, He H, \Xu J, Yang B,Liu D (2025) Proteomic Analysis of Diabetic Foot Skin Highlights Neutrophil Dysfunction and Identifies Potential Therapeutic Targets MDL-28170 and Amlexanox. JSM Foot Ankle 6(1): 1055.
INTRODUCTION
Diabetic foot (DF) and its associated lower limb complications have emerged as a formidable public health challenge for the global diabetic population, posing significant morbidity and mortality risks [1,2]. Current estimates suggest that between 40 to 60 million individuals worldwide are affected by DF, with approximately 15% of diabetic patients classified as high-risk and 5% actively suffering from the debilitating effects of this condition. Diabetic foot ulcers (DFUs), a severe manifestation of DF, are particularly concerning due to their potential to progress to limb amputation, thereby drastically diminishing the quality of life and contributing to premature mortality [3 6]. The psychological and physiological burden borne by patients and their families is profound, underscoring the urgency of addressing this escalating health crisis [7,8]. The pathogenesis of DF is a multifactorial and multistage process characterized by the intricate interplay of external and internal factors [9,10]. External factors such as colonization and infection of wounds, prolonged pressure, and recurrent trauma exacerbate inflammatory responses, significantly delaying the healing process. The cumulative impact of these factors can perpetuate and intensify tissue damage [10-12]. Internally, the presence of neuropathy and inadequate blood supply further disrupts the metabolic states and functions of various cell types critical to wound healing, complicating and often impeding the normal physiological repair processes [13 15]. These intrinsic factors not only hinder recovery but also exacerbate the progression of the disease. In non-healing DFUs, elevated levels of specific biomarkers, including elastase, neutrophil gelatinase associated lipocalin (Matrix Metalloproteinase-9, MMP 9), and elastase, are commonly associated with infection and delayed healing [16,17]. The overexpression of these biomarkers can lead to the degradation of tissue structure, thereby obstructing wound closure. Moreover, macrophage-derived chemokine (MDC), Th2-type CC chemokines, thymus and activation-regulated chemokine (TARC), as well as clusterin and glycoprotein 4, have been identified as potential prognostic biomarkers for DFU healing [18-20]. These factors play crucial roles in regulating immune responses and promoting wound repair. Notably, the expression of certain anti-angiogenic proteins, such as COL15A1, COL18A1, collagen, and tenascin, is upregulated in chronic DFU wounds [21 23]. The increase in these proteins may contribute to the pathology of chronic DFU by inhibiting angiogenesis, thereby hindering wound healing and offering new perspectives for the treatment of diabetic foot [24,25]. In this study, we conducted an integrated analysis of proteomic data from DF samples and publicly available gene expression profiles to uncover the abnormal enrichment of neutrophil degranulation and neutrophil extracellular trap (NET) signalling in DF tissues. These findings not only deepen our understanding of the pathological mechanisms underlying DF but also provide critical insights for identifying potential drug targets. Through further research, we aim to develop novel therapeutic strategies that improve patient outcomes, reduce the incidence and progression of DFUs, and ultimately enhance the quality of life for diabetic patients. The objective of this research is to explore the pathogenesis of diabetic foot comprehensively, identify key biomarkers, and leverage these discoveries to develop new treatment strategies. By employing an interdisciplinary approach that integrates proteomic and gene expression analyses, we hope to gain a more holistic understanding of the complexities of diabetic foot and offer more effective therapeutic solutions for patients.
MATERIALS AND METHODS
Sample Collection and Preparation
Diabetic foot skin samples (n=3) and control non-diabetic foot skin samples (n=3) were obtained from Zhongda Hospital, Southeast University, Nanjing, China, following approval by the institutional ethics committee (Ethics approval ID:2018ZDSYLL132-P01). The collected tissue samples were immediately snap-frozen in liquid nitrogen and subsequently ground into fine cell powder using a cryogenic grinder. The powdered samples were then transferred to 5 mL centrifuge tubes, and four volumes of lysis buffer (8 M urea, 1% protease inhibitor cocktail) were added. The samples were subjected to sonication on ice using a high-intensity ultrasonic processor (Scientz) for three cycles to ensure efficient lysis. Following sonication, the lysates were centrifuged at 12,000 g for 10 minutes at 4 °C to remove any remaining debris. The supernatant containing the solubilized proteins was carefully collected, and protein concentration was determined using a bicinchoninic acid (BCA) assay kit as per the manufacturer’s instructions.
LC-MS/MS
Analysis Proteomic profiling was conducted using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Tryptic peptides, prepared from the protein extracts, were dissolved in 0.1% formic acid (solvent A) and loaded onto a homemade reversed-phase analytical column (15 cm in length, 75 μm inner diameter). A gradient elution was performed on an EASY-nLC 1000 UPLC system, starting with 6% solvent B (0.1% formic acid in 98% acetonitrile) and increasing to 23% over 26 minutes, followed by an increase to 35% over 8 minutes, and finally to 80% over 3 minutes, maintaining this level for an additional 3 minutes at a constant flow rate of 400 nL/min. The separated peptides were then introduced into a Q Exactive™ Plus mass spectrometer (Thermo Fisher Scientific) via a nano electrospray ionization (NSI) source operating at a voltage of 2.0 kV. Full-scan MS spectra were acquired in the m/z range of 350 to 1800 at a resolution of 70,000 in the Orbitrap analyzer. The most abundant precursor ions were selected for fragmentation using higher-energy collisional dissociation (HCD) with a normalized collision energy (NCE) setting of 28. Fragment ions were detected in the Orbitrap at a resolution of 17,500. The mass spectrometer operated in a data-dependent acquisition mode, alternating between a full MS scan followed by 20 MS/MS scans with a dynamic exclusion time of 15 seconds. The automatic gain control (AGC) target was set to 5E4 with a maximum injection time of 200 ms, and the fixed first mass was set at 100 m/z.
Proteomics Data Processing
The mass spectrometry analysis generated a total of 322,586 MS/MS spectra. These spectra were searched against the theoretical protein database using a proteomics search engine, resulting in the identification of 51,020 high-confidence spectra with a utilization rate of 15.8%. From these, 26,624 unique peptides were identified, with 25,501 being unique peptide sequences. Overall, 4,202 proteins were identified, of which 2,821 could be quantified (quantifiable proteins were those with quantification information available in at least one comparison group). Detailed experimental data statistics are summarized in Table, and raw data files are available in the accompanying dataset “S8190TQ/2-Basic_analysis/ MS_identified_information.xlsx.”
Public Data Acquisition
Publicly available gene expression datasets were utilized to complement the proteomic findings. Data were retrieved from the Gene Expression Omnibus (GEO) under the accession numbers GSE68183 [26,27], which include transcriptomic profiles from diabetic foot ulcer (DFU) and non-diabetic foot skin samples.
Differential Expression Analysis
Quantitative analysis of differentially expressed proteins was performed using data from three biological replicates per sample. For each protein, the average of the quantification values from the three replicates was calculated. Differential expression between diabetic foot and non-diabetic foot samples was assessed by computing the ratio of these average values. To determine the statistical significance of the differential expression, the relative quantification values were log2-transformed to approximate a normal distribution. A two-tailed Welch’s t-test [28,29], was then applied to calculate the p-value for each protein. Proteins with a p-value < 0.05 and a log2 transformed fold change greater than 1 or less than -1 were considered significantly upregulated or downregulated, respectively.
Gene Ontology Enrichment Analysis
Differentially expressed proteins were annotated and categorized according to Gene Ontology [30], (GO) terms across three main domains: biological process, cellular component, and molecular function. Enrichment analysis for each category was performed using a two-tailed Fisher’s exact test, comparing the frequency of each GO term in the set of differentially expressed proteins against its frequency in the background set comprising all identified proteins. To account for multiple hypothesis testing, p-values were adjusted using the Benjamini-Hochberg procedure. GO terms with an adjusted p-value of less than 0.05 were considered significantly enriched. This analysis provided insights into the biological processes, cellular structures, and molecular functions most perturbed in diabetic foot pathology.
Pathway Enrichment Analysis
Pathway enrichment analysis was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [31], to identify pathways significantly associated with the differentially expressed proteins. A two-tailed Fisher’s exact test was employed to assess the enrichment of these proteins within known KEGG pathways relative to all identified proteins. Pathways with a Benjamini-Hochberg corrected p-value of less than 0.05 were deemed significantly enriched.
Metascape Functional Enrichment Analysis
Metascape (http://metascape.org) was utilized for comprehensive functional enrichment analysis to identify key biological processes, pathways, and GO terms associated with the gene set of interest [32]. The analysis involved several key steps: (1) Submission of the gene list to Metascape, which integrates multiple biological databases, including GO, KEGG, Reactome [33], and BioCarta [34], for broad coverage of potential biological functions. (2) Comparison of the input gene list against a background set, typically the entire genome of the studied organism, to identify significantly enriched GO terms and pathways. (3) Calculation of p-values using the hypergeometric test, followed by adjustment for multiple comparisons using the Benjamini-Hochberg procedure to control the false discovery rate (FDR). GO terms and pathways with an FDR-adjusted p-value of less than 0.05 were considered significant. (4) Visualization of the enrichment results through bar graphs, cluster heatmaps, and network diagrams, facilitating the interpretation of interconnected biological themes. This analysis allowed for the generation of a comprehensive overview of the biological functions implicated in diabetic foot ulceration.
Connectivity Map (CMap) Drug Enrichment and Molecular Docking Analysis
To identify potential therapeutic compounds for diabetic foot ulcers, the Connectivity Map (CMap) was employed [35]. The gene expression signatures derived from the proteomic and transcriptomic data were queried against the CMap database, which houses an extensive repository of gene expression profiles induced by various drugs. This analysis was aimed at identifying compounds that could potentially reverse or mimic the disease-related gene expression changes. The compounds were ranked based on their connectivity scores, with higher scores indicating stronger correlations with the input gene signature. The top candidate drugs were then selected for molecular docking studies to evaluate their binding affinity and interactions with target proteins. Molecular docking was performed using AutoDock Vina [36], a widely recognized tool for this purpose. The three-dimensional structures of the target proteins and drug molecules were retrieved from the Protein Data Bank (PDB [37], and PubChem [38], respectively. Docking simulations were conducted using standard parameters, and the resulting drug-target complexes were analyzed to determine their binding affinities and key molecular interactions.Complexes exhibiting the lowest binding energies were prioritized for further validation and experimental testing.
Statistical Analysis
All statistical analyses were performed using R software and other specialized tools where appropriate. The significance of GO terms, pathways, and drug-target interactions was rigorously tested using established statistical methods, ensuring robust and reproducible results. The findings were validated through multiple iterations of analysis, with p-values adjusted to control for false discoveries across the multiple comparisons conducted in this study.
RESULTS
Proteomic Differences Between Diabetic Foot Skin (DFS) and Non-Diabetic Foot Skin (NFS)
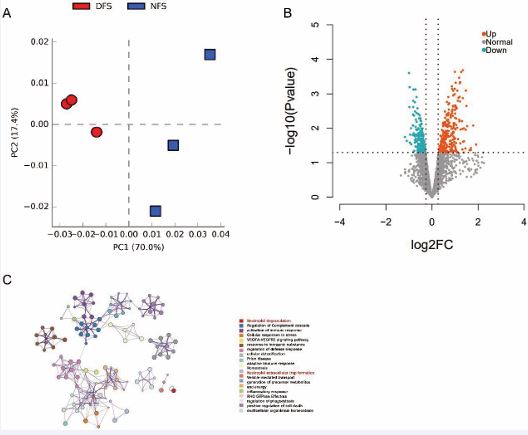
Principal component analysis (PCA) of the proteomic data from diabetic foot skin (DFS) and non-diabetic foot skin (NFS) samples revealed distinct clustering, with the first principal component (PC1) accounting for 70% of the variance and the second principal component (PC2) explaining 17.4% of the variance (Figure 1A). This significant separation indicates profound proteomic differences between DFS and NFS tissues. To identify proteins with significant differential expression, a threshold of log2 fold change (log2FC) > 1 and a Welch t-test p-value < 0.05 was applied. This analysis identified 449 proteins with significant expression differences, including 244 proteins that were upregulated and 205 that were downregulated in DFS compared to NFS (Figure 1B). Functional enrichment analysis using Metascape revealed that these differentially expressed proteins were significantly enriched in biological processes related to neutrophil degranulation, complement cascade regulation, and immune response activation (Figure 1C). Given the well-established role of neutrophils in the pathogenesis of diabetic wounds, particularly in promoting inflammation and delayed healing, we focused further analysis on neutrophil degranulation and neutrophil extracellular trap (NET) formation pathways [39,40]. NETs have been implicated as key drivers of diabetic complications, including impaired wound healing and diabetic retinopathy [41]. Elevated glucose levels can induce NET-mediated cell death, a natural defense mechanism that, when overactivated, contributes to thrombosis, inflammation, and endothelial dysfunction [42,43].
Figure 1 Principal Component Analysis (PCA) and Differential Protein Expression in Diabetic Foot Skin (DFS) vs. Non-Diabetic Foot Skin (NFS)
Figure 1A: Principal Component Analysis (PCA) of proteomic data from diabetic foot skin (DFS) and non-diabetic foot skin (NFS) samples. The PCA plot demonstrates distinct clustering of the DFS and NFS samples, with the first principal component (PC1) accounting for 70% of the variance and the second principal component (PC2) explaining 17.4% of the variance. The clear separation between DFS and NFS samples indicates significant differences in their proteomic profiles.
Figure 1B: Volcano plot illustrating the differential expression of proteins between DFS and NFS samples. Proteins with a log2 fold change greater than 1 or less than -1 and a Welch t-test p-value < 0.05 are considered significantly differentially expressed. A total of 449 proteins were identified as differentially expressed, with 244 proteins upregulated and 205 proteins downregulated in DFS compared to NFS.
Figure 1C: Functional enrichment analysis of differentially expressed proteins using Metascape. The bar graph shows the top enriched biological processes, with a focus on neutrophil degranulation, complement cascade regulation, and immune response activation. These processes are highly relevant to the pathogenesis of diabetic foot, particularly in the context of inflammation and impaired wound healing.
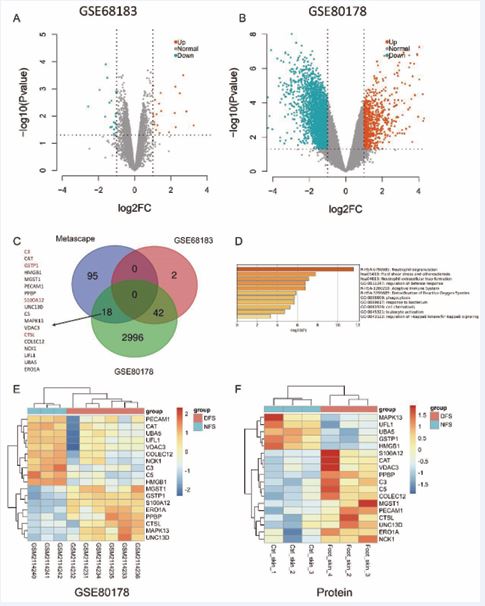
Association of Neutrophil Degranulation and NET Formation with Diabetic Foot Pathogenesis To further investigate the involvement of neutrophil-related functions in diabetic foot pathology, we conducted differential expression analysis using public datasets GSE80178 (comprising six diabetic foot ulcer samples and three non-diabetic foot skin samples) and GSE68183 (comprising three diabetic foot skin samples and three non-diabetic foot skin samples). In the GSE80178 dataset, 741 genes were significantly upregulated, and 2,424 genes were significantly downregulated. Similarly, in the GSE68183 dataset, 21 genes were upregulated, and 23 were downregulated (Figure 2A,B). A Venn diagram analysis of the differentially expressed proteins associated with neutrophil degranulation and NET formation identified 18 common targets implicated in diabetic foot pathology (Figure 2C). Notably, four of these targets were upregulated by more than 4-fold. Visualization of these 18 genes in the GSE80178 dataset revealed that 10 genes (PECAM1, CAT, UBA5, UFL1, VDAC3, COLEC12, NCK1, C3, C5, HMGB1) were significantly upregulated in non-diabetic foot samples, while eight genes (MGST1, GSTP1, S100A12, ERO1A, PPBP, CTSL, MAPK13, UNC13D) were upregulated in diabetic foot samples(Figure 2D). Proteomic data corroborated these findings, showing significant upregulation of MAPK13, UFL1, UBA5, GSTP1, and HMGB1 in NFS, while S100A12, CAT, VDAC3, PPBP, C3, C5, COLEC12, MGST1, PECAM1, CTSL, UNC13D, ERO1A, and NCK1 were significantly upregulated in DFS (Figure 2E). Metascape functional enrichment analysis of these 18 intersecting proteins highlighted their strong association with neutrophil degranulation and NET formation, reinforcing their potential role in the pathogenesis of diabetic foot ulcers (Figure 2F).
Figure 2 Differential Gene Expression Analysis and Identification of Key Proteins in Diabetic Foot Pathogenesis
Figure 2A: Volcano plot of differentially expressed genes from the GSE68183 dataset. This plot illustrates the distribution of gene expression changes, highlighting the significantly upregulated and downregulated genes between diabetic foot samples and non-diabetic controls.
Figure 2B: Volcano plot of differentially expressed genes from the GSE80178 dataset. Similar to Figure 2A, this plot shows the significant gene expression changes, emphasizing the genes most affected in diabetic foot pathology.
Figure 2C: Venn diagram illustrating the overlap of differentially expressed genes from two public datasets, GSE80178 and GSE68183, and our differentially expressed proteins associated with neutrophil-related signaling pathways. The diagram shows the number of upregulated and downregulated genes in each dataset, with 18 key genes shared across these datasets, implicating their critical role in the pathogenesis of diabetic foot.
Figure 2D, E: Heatmaps displaying the expression levels of the 18 key genes in the GSE80178 dataset and in our proteomic quantitative cohort. These heatmaps highlight significant upregulation of genes involved in neutrophil degranulation and related processes (e.g., S100A12, CTSL) in diabetic foot samples, reinforcing their potential contribution to disease pathogenesis.
Figure 2F: Metascape functional enrichment analysis of the 18 intersecting proteins. The analysis identifies key biological processes associated with these proteins, such as neutrophil degranulation, immune response activation, and NET formation. These findings emphasize the central role of these processes in diabetic foot pathology, suggesting their importance as potential therapeutic targets.
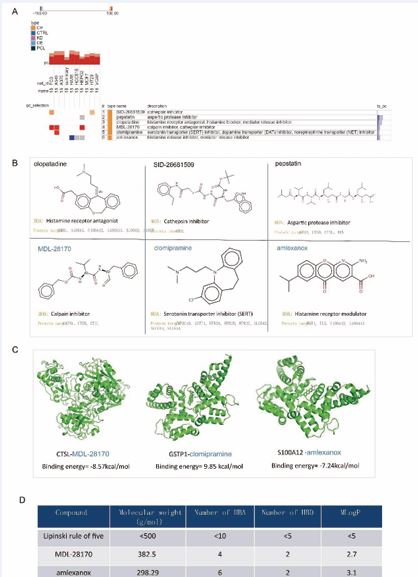
Identification of Potential Therapeutic Compounds for Diabetic Foot Ulcers To identify potential therapeutic agents targeting the identified key proteins, we utilized the Connectivity Map (CMap) database. We screened for drugs targeting the 18 significant proteins and identified six candidate compounds. Based on their connectivity scores (≥95 or ≤-95 across various cell lines), three compounds—MDL 28170, clomipramine, and amlexanox—were selected for further analysis (Figure 3A,B).
Figure 3 Identification of Potential Therapeutic Compounds and Molecular Docking Analysis
Figure 3A: Connectivity Map (CMap) drug enrichment analysis identifying potential therapeutic compounds targeting the 18 key proteins implicated in diabetic foot pathology. The bar graph displays the top-ranked compounds based on their connectivity scores, with MDL-28170, clomipramine, and amlexanox emerging as the most promising candidates.
Figure 3B: Molecular docking results for MDL-28170 and amlexanox with their respective target proteins. The table summarizes the binding energies and key interactions for the top drug-target pairs, with lower binding energies indicating stronger affinities. MDL-28170 and amlexanox show strong binding to CTSL and S100A12, respectively, suggesting potential therapeutic efficacy.
Figure 3C: 3D models of the molecular docking interactions between MDL-28170/CTSL and amlexanox/S100A12. The models depict the drugs binding to the active sites of their target proteins, with hydrogen bonds and hydrophobic interactions stabilizing the complexes. These interactions support the potential use of these compounds in therapeutic interventions for diabetic foot.
Figure 3D: Evaluation of the selected compounds (MDL-28170, clomipramine, and amlexanox) for drug-likeness using Lipinski’s rule of five. The table lists the physicochemical properties of each compound, including molecular weight, hydrogen bond donors and acceptors, and LogP values. All compounds meet the criteria for oral bioavailability, suggesting favorable pharmacokinetic profiles and potential for further drug development.
Molecular docking studies were performed using AutoDock Vina to evaluate the binding affinity of these drugs with their respective target proteins. The docking results demonstrated low binding energies for the CTSL-MDL-28170 and S100A12 amlexanox interactions, indicating strong binding affinity and potential therapeutic efficacy (Figure 3C). MDL 28170, a selective inhibitor of calpain and cathepsin B, has been shown to penetrate the blood-brain barrier and inhibit cysteine protease activity in various rodent models, offering neuroprotective benefits [44,45]. Amlexanox, a pyridochromene derivative with anti-inflammatory, anti ulcer, and non-steroidal anti-inflammatory properties, has been clinically effective in treating atopic diseases, particularly allergic asthma and rhinitis [46]. Amlexanox has also shown promise in enhancing insulin sensitivity and glucose metabolism in diabetic models [47,48]. The selected compounds were further evaluated for their potential as drug candidates by applying Lipinski’s rule of five, a widely recognized set of criteria used to assess the drug-likeness of a compound [49]. This rule posits that an orally active drug is more likely to be absorbed and permeable if it meets certain criteria: it should have no more than five hydrogen bond donors, no more than ten hydrogen bond acceptors, a molecular weight under 500 Daltons, and a LogP (partition coefficient between octanol and water) not greater than five. These criteria are essential for ensuring that a compound possesses the necessary pharmacokinetic properties for oral bioavailability, such as adequate absorption, distribution, metabolism, and excretion (ADME) profiles. Upon thorough evaluation, all selected compounds were found to fully comply with Lipinski’s rule of five, indicating that they possess favorable physicochemical properties for oral administration (Figure 3D). No violations of the rule were observed, suggesting that these compounds are likely to exhibit good bioavailability and pharmacokinetic stability in the body. This compliance with the rule of five strengthens the case for these compounds as viable candidates for further drug development, as it implies a higher probability of success in subsequent stages of preclinical and clinical testing. The absence of any rule violations also underscores the potential of these compounds to move forward in the drug discovery pipeline with reduced risk of encountering ADME-related issues during development.
DISCUSSION
Diabetic foot (DF) has emerged as a critical complication among the growing global population of diabetes patients, presenting formidable challenges in both clinical management and public health [1,50]. As the incidence of diabetes continues to rise, so too does the prevalence of DF and its associated complications, including diabetic foot ulcers (DFUs), which can lead to significant morbidity, amputations, and even premature mortality [51,52]. The complex pathophysiology of DF involves a multifaceted interplay of systemic metabolic dysfunction, local tissue ischemia, neuropathy, and persistent infections, all contributing to the chronicity and severity of the condition [53-56].In this study, we performed a comprehensive proteomic analysis comparing DF skin tissue with non-diabetic foot (NDF) skin tissue, revealing a marked disparity in protein expression profiles between the two groups. Our analysis identified 449 differentially expressed proteins, with a significant focus on neutrophil-related functions, coagulation, and angiogenesis. These findings are consistent with the established role of neutrophils in the pathogenesis of diabetic complications, particularly their contribution to the impaired wound healing observed in DFUs. Neutrophil extracellular traps (NETs), in particular, have been implicated in exacerbating inflammation and thrombosis, which are hallmark features of diabetic vascular complications. Previous studies have highlighted the pro-thrombotic and pro-inflammatory effects of NETs in diabetic wounds, aligning with our observations of enriched neutrophil degranulation and NET formation pathways in DF tissues. Moreover, by integrating public gene expression datasets, we identified 18 key proteins whose differential expression was corroborated at the transcriptomic level. Notably, the functional enrichment of these proteins also centered on neutrophil degranulation and platelet activation, further underscoring the significance of immune dysregulation in DF pathology. Our findings of elevated expression levels of proteins such as S100A12, GSTP1, and CTSL in DF tissues suggest their potential as biomarkers for disease severity and as therapeutic targets. The selection of MDL-28170 and amlexanox as candidate drugs through Connectivity Map (CMap) analysis and subsequent molecular docking provides a promising avenue for targeted therapy. MDL-28170, a calpain and cathepsin B inhibitor, has shown neuroprotective effects in diabetic models [57], while amlexanox, an anti inflammatory and anti-ulcer agent, may improve wound healing by modulating glucose metabolism and immune responses [58]. Our study contributes to the growing body of literature on DF by providing a proteomic perspective that complements existing genomic data, thereby offering a more holistic understanding of the disease mechanisms. However, it is important to acknowledge certain limitations in our research. The relatively small sample size may limit the generalizability of our findings, and the proteomic data require further validation in larger, independent cohorts. Additionally, while our molecular docking results are promising, in vivo studies are necessary to confirm the therapeutic efficacy of the identified compounds In summary, our study underscores the critical role of neutrophil dysfunction in the pathogenesis of DF and highlights potential therapeutic targets for improving patient outcomes. Future research should focus on validating these findings in clinical settings and exploring the therapeutic potential of the identified compounds to mitigate the burden of diabetic foot complications.
Data availability statement
The proteomics data generated and analyzed during this study are available upon reasonable request from the corresponding authors. The transcriptomics data utilized in this study are available through the GEO database, with specific access details provided in the Methods section.
Acknowledgments
We acknowledge the GEO database for providing publicly accessible transcriptomics data, which greatly facilitated the research conducted in this study.
Conflict of interest
The authors declare that there are no commercial or financial relationships that could be construed as a potential conflict of interest in this study.
Ethics statement
The ethics approval and consent procedures for this study are detailed in the Methods section.
Author contributions
Han Yin and Hua He contributed equally to this work. Han Yin (First Author) was primarily responsible for the conceptualization, methodology, data collection, and manuscript drafting. Hua He assisted with the data analysis and provided critical feedback on the manuscript. Juan Xu contributed to the interpretation of the results and assisted with manuscript editing. Delin Liu (Corresponding Author) supervised the entire project, oversaw the study design, and provided important guidance throughout the research process. Bingquan Yang (Co-corresponding Author) contributed to the overall supervision and provided expertise in data analysis and interpretation.
REFERENCES
- Guan H, Wang Y, Niu P, Zhang Y, Zhang Y, Miao R, et al. The role of machine learning in advancing diabetic foot: a review. Front Endocrinol (Lausanne). 2024; 15: 1325434.
- Zhang HM, Yang ML, Xi JZ, Yang GY, Wu QN. Mesenchymal stem cells- based drug delivery systems for diabetic foot ulcer: A review. World J Diabetes. 2023; 14: 1585.
- Matijevi? T, Talapko J, Meštrovi? T, Matijevi? M, Eri? S, Eri? I, et al. Understanding the multifaceted etiopathogenesis of foot complications in individuals with diabetes. World J Clin Cases. 2023; 11: 1669-1683.
- Armstrong DG, Boulton AJM, Bus SA. Diabetic Foot Ulcers and Their Recurrence. N Engl J Med. 2017; 376: 2367-2375.
- Singh N, Armstrong DG, Lipsky BA. Preventing foot ulcers in patients with diabetes. JAMA. 2005; 293: 217-228.
- IDF DA. International diabetes federation. Diabetes Research and Clinical Practice. 2021; 102.
- Hogg FR, Peach G, Price P, Thompson MM, Hinchliffe RJ. Measures of health-related quality of life in diabetes-related foot disease: a systematic review. Diabetologia. 2012; 55: 552-565.
- Pickwell KM, Siersma VD, Kars M, Holstein PE, Schaper NC; Eurodiale consortium. Diabetic foot disease: impact of ulcer location on ulcer healing. Diabetes Metab Res Rev. 2013; 29: 377-383.
- Boulton AJ, Vileikyte L, Ragnarson-Tennvall G, Apelqvist J. The global burden of diabetic foot disease. Lancet. 2005; 366: 1719-1724.
- Armstrong DG, Lipsky BA. Diabetic foot infections: stepwise medical and surgical management. Int Wound J. 2004; 1: 123-132.
- Bakker K, Apelqvist J, Schaper NC; International Working Group on Diabetic Foot Editorial Board. Practical guidelines on the management and prevention of the diabetic foot 2011. Diabetes Metab Res Rev. 2012; 28: 225-231.
- Lavery LA, Armstrong DG, Harkless LB. Classification of diabetic foot wounds. J Foot Ankle Surg. 1996; 35: 528-531.
- Burgess JL, Wyant WA, Abdo Abujamra B, Kirsner RS, Jozic I. Diabetic Wound-Healing Science. Medicina (Kaunas). 2021; 57: 1072.
- Boulton AJ, Armstrong DG, Albert SF, Frykberg RG, Hellman R, Kirkman MS, et al. Comprehensive foot examination and risk assessment: a report of the task force of the foot care interest group of the American Diabetes Association, with endorsement by the American Association of Clinical Endocrinologists. Diabetes Care. 2008; 31: 1679-1685.
- Lipsky BA, Berendt AR, Cornia PB, Pile JC, Peters EJ, Armstrong DG, et al. Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin Infect Dis. 2012; 54: e132-e173.
- Lazaro JL, Izzo V, Meaume S, Davies AH, Lobmann R, Uccioli L. Elevated levels of matrix metalloproteinases and chronic wound healing: an updated review of clinical evidence. J Wound Care. 2016; 25: 277-287.
- Tavakoli F, Faramarzi M, Salimnezhad S, Jafari B, Eslami H, Mohammad PourTabrizi B. Comparing the activity level of salivary matrix metalloproteinase-8 in patients with diabetes and moderate to severe chronic generalized periodontitis. Clin Exp Dent Res. 2024; 10: e865.
- Rayment EA, Upton Z. Finding the culprit: a review of the influences of proteases on the chronic wound environment. Int J Low Extrem Wounds. 2009; 8: 19-27.
- Wysocki AB, Staiano-Coico L, Grinnell F. Wound fluid from chronic leg ulcers contains elevated levels of metalloproteinases MMP-2 and MMP-9. J Invest Dermatol. 1993; 101: 64-68.
- Galkowska H, Olszewski WL, Wojewodzka U, Rosinski G, Karnafel W. Neurogenic factors in the impaired healing of diabetic foot ulcers. J Surg Res. 2006; 134: 252-258.
- O’Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, et al. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997; 88: 277-285.
- Wight TN, Kinsella MG, Qwarnström EE. The role of proteoglycans in cell adhesion, migration and proliferation. Curr Opin Cell Biol. 1992; 4: 793-801.
- Di Lullo GA, Sweeney SM, Korkko J, Ala-Kokko L, San Antonio JD. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J Biol Chem. 2002; 277: 4223-4231.
- Falanga V. Wound healing and its impairment in the diabetic foot. Lancet. 2000; 366: 1736-1743.
- Veves A, Falanga V, Armstrong DG, Sabolinski ML. Apligraf Diabetic Foot Ulcer Study. Graftskin, a human skin equivalent, is effective in the management of noninfected neuropathic diabetic foot ulcers: a prospective randomized multicenter clinical trial. Diabetes Care. 2001; 24: 290-295.
- Ramirez HA, Liang L, Pastar I, Rosa AM, Stojadinovic O, Zwick TG, et al. Comparative Genomic, MicroRNA, and Tissue Analyses Reveal Subtle Differences between Non-Diabetic and Diabetic Foot Skin. PLoS One. 2015; 10: e0137133.
- Amirez HA, Pastar I, Jozic I, Stojadinovic O, Stone RC, Ojeh N, et al. Staphylococcus aureus Triggers Induction of miR-15B-5P to Diminish DNA Repair and Deregulate Inflammatory Response in Diabetic Foot Ulcers. J Invest Dermatol. 2018; 138: 1187-1196.
- Delacre M, Lakens D, Leys C. Why psychologists should by default use Welch’s t-test instead of Student’s t-test. Int Rev Social Psychol. 2017; 30: 92-101.
- Zimmerman DW, Zumbo BD. Rank transformations and the power of the Student t test and Welch t’test for non-normal populations with unequal variances. Canadian J Exp Psychol Revue canadienne de psychologie expérimentale. 1993; 47: 523.
- Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015; 43(Database issue): D1049-D1056.
- Lu S, Wang H, Zhang J. Identification of uveitis-associated functions based on the feature selection analysis of gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment scores. Front Mol Neurosci. 2022; 15: 1007352.
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019; 10: 1523.
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018; 46(D1): D649-D655.
- Nishimura D. BioCarta. Biotech Software & Internet Report: The Computer Software J Scient. 2001; 2: 117-120.
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007; 7: 54-60.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010; 31: 45561.
- Burley SK, Berman HM, Kleywegt GJ, Markley JL, Nakamura H, VelankarS. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol Biol. 2017; 1607: 627-641.
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019; 47: D1102-D1109.
- Yang S, Gu Z, Lu C, Zhang T, Guo X, Xue G, et al. Neutrophil Extracellular Traps Are Markers of Wound Healing Impairment in Patients with Diabetic Foot Ulcers Treated in a Multidisciplinary Setting. Adv Wound Care (New Rochelle). 2020; 9: 16-27.
- Zhao R, Liang H, Clarke E, Jackson C, Xue M. Inflammation in Chronic Wounds. Int J Mol Sci. 2016; 17: 2085.
- Berezin A. Neutrophil extracellular traps: The core player in vascular complications of diabetes mellitus. Diabetes Metab Syndr. 2019; 13: 3017-3023.
- Hisada Y, Grover SP, Maqsood A, Houston R, Ay C, Noubouossie DF, et al. Neutrophils and neutrophil extracellular traps enhance venous thrombosis in mice bearing human pancreatic tumors. Haematologica. 2020; 105: 218-225.
- Zhou Y, Xu Z, Liu Z. Impact of Neutrophil Extracellular Traps on Thrombosis Formation: New Findings and Future Perspective. Front Cell Infect Microbiol. 2022; 12: 910908.
- Thompson SN, Carrico KM, Mustafa AG, Bains M, Hall ED. A pharmacological analysis of the neuroprotective efficacy of the brain- and cell-permeable calpain inhibitor MDL-28170 in the mouse controlled cortical impact traumatic brain injury model. J Neurotrauma. 2010; 27: 223343.
- Carragher NO, Frame MC. Calpain: a role in cell transformation and migration. Int J Biochem Cell Biol. 2002; 34: 1539-1543.
- Dosanjh A, Won CY. Amlexanox: A Novel Therapeutic for Atopic, Metabolic, and Inflammatory Disease. Yale J Biol Med. 2020; 93: 759- 763.
- Bell J. Amlexanox for the treatment of recurrent aphthous ulcers. Clin Drug Investig. 2005; 25: 555-566.
- Reilly SM, Chiang SH, Decker SJ, Chang L, Uhm M, Larsen MJ, et al. An inhibitor of the protein kinases TBK1 and IKK-? improves obesity- related metabolic dysfunctions in mice. Nat Med. 2013; 19: 313-321.
- Zhang MQ, Wilkinson B. Drug discovery beyond the ‘rule-of-five’. Curr Opin Biotechnol. 2007; 18: 478-488.
- Senneville É, Albalawi Z, van Asten SA, Abbas ZG, Allison G, Aragón- Sánchez J, et al. Diagnosis of infection in the foot of patients with diabetes: A systematic review. Diabetes Metab Res Rev. 2024; 40: e3723.
- McDermott K, Fang M, Boulton AJM, Selvin E, Hicks CW. Etiology, Epidemiology, and Disparities in the Burden of Diabetic Foot Ulcers. Diabetes Care. 2023; 46: 209-221.
- Peter-Riesch B. The Diabetic Foot: The Never-Ending Challenge. Endocr Dev. 2016; 31: 134.
- Kim J. The pathophysiology of diabetic foot: a narrative review. J Yeungnam Med Sci. 2023; 40: 328-334.
- örneskog G. Why critical limb ischemia criteria are not applicable to diabetic foot and what the consequences are. Scand J Surg. 2012; 101: 114-118.
- Pitocco D, Spanu T, Di Leo M, Vitiello R, Rizzi A, Tartaglione L, et al. Eur Rev Med Pharmacol Sci. 2019; 23: 26-37.
- Uçkay I, Gariani K, Pataky Z, Lipsky BA. Diabetic foot infections: state- of-the-art. Diabetes Obes Metab. 2014; 16: 305-316.
- Yuen P, Wang KK. Calpain inhibitors: novel neuroprotectants and potential anticataract agents. Drugs of the Future. 1998; 23: 741-750.
- Bailly C. The potential value of amlexanox in the treatment of cancer: Molecular targets and therapeutic perspectives. Biochem Pharmacol. 2022; 197: 114895.