A Rapid Cost-Effective Detection of Toxigenic Clostridioides difficile from Diarrheal Stools and Presumptive Identification of NAP1 Strain using Multiplex Loop-Mediated Isothermal Amplification (LAMP)
- 1. Department of Pathology and Molecular Medicine, McMaster University, Canada
- 2. Microbiology Section, Hamilton Regional laboratory Medicine Program, St. Joseph’s Healthcare, Hamilton, Canada
ABSTRACT
This study describes the development of a cost-effective, multiplex Loop-Mediated Isothermal Amplification (LAMP) method for detection of toxigenic Clostridioides difficile from diarrheal stools and presumptive identification of the NAP-1 strain. The diagnostic values for the new method were as follows: 100.0% specificity, 95.0% sensitivity, 100% positive predictive value, and 97.0% negative predictive value as compared to real-time PCR. The estimated cost per test is Cdn$ 3.75 and which is significantly less than commercial assays. The average turn-around-time from set-up to detection is 1.5 h. The LAMP method described here is a cost-effective, quick and highly sensitive test which can be implemented in a clinical laboratory to assist clinicians in establishing the diagnosis of CDI and to indirectly determine the presence of the hypervirulent epidemic binary toxin producing NAP1 strain.
CITATION
Jayaratne P, Lee C (2022) A Rapid Cost-Effective Detection of Toxigenic Clostridioides dificile from Diarrheal Stools and Presumptive Iden- tification of NAP1 Strain using Multiplex Loop-Mediated Isothermal Amplification (LAMP). JSM Gastroenterol Hepatol 9(2): 1110.
INTRODUCTION
Clostridioides difficile-associated disease (CDAD) is a leading cause of nosocomial diarrhea in adults. Symptoms can range from mild, self-limiting diarrhea to pseudomembraneous colitis to fulminant toxic megacolon, resulting in adverse outcomes. During the past decade there has been a substantial increase in the number of cases and an escalating rate of serious disease with a four-fold increase in mortality [1]. There has been significant morbidity and mortality related to C. difficile infection (CDI) due to the presence of the hypervirulent strain (NAP1) associated with unregulated production of toxin [1]. NAP1 has been responsible for a number of outbreaks in many countries [2]. Therefore, rapid and accurate reporting of C. difficile is essential for improving patient outcomes and minimizing hospital-acquired disease. Now rapid PCR-based commercial and in-house methods that approach the sensitivity and specificity of culture-toxin which is the gold standard in diagnosing CDAD are available.
However, the PCR methods require nucleic acid purification and are therefore too expensive for routine testing. The only other commercially available isothermal nucleic acid-based amplification test (illumigene®, Meridian Biosciences Inc) that does not require DNA purification is also expensive and will not identify the NAP1 strain.
Here we describe an inexpensive, multiplex real-time Loop-Mediated Isothermal Amplification method (LAMP) to detect toxigenic C. difficile and to presumptively identify the hypervirulent NAP1 strain directly from diarrheal stools without nucleic acid purification.
Patient Population
Five-hundred and eighteen unformed stool specimens from adults with suspected CDI submitted to the microbiology laboratory at the Hamilton Health Sciences and St. Joseph’s Healthcare, Hamilton, Ontario, Canada were tested. Evaluation of these specimens was also done by the current routine Real- Time PCR (that has been previously validated with 2 commercial assays) for the detection of C. difficile toxin A and/or B according to the standard operational procedure. The in-house real-time PCR method detected the tcdC (surrogate marker for tcdA/tcdB toxin genes), and cdtA (binary toxin gene). Genes were amplified using
the QuantiTectTM Multiplex PCR Kit (Qiagen Inc.) and detected by TaqMan probes. Both methods were done in a Rotor-Gene 6500 (Qiagen Inc.). For this study, the gold standard was defined as 100% concordance between tests.
DNA Extraction
For the in-house Real-Time PCR method 100ul of thawed stool samples were emulsified in 900ul of Sputum Liquefying Solution (SLS; Copan, Italy). and 200ul of the supernatant was extracted using the easyMAG™ (bioMérieux) automated nucleic acid extractor according to the manufacturer’s instructions. Purified DNA was collected in 55ul of elution buffer.
For LAMP, DNA was extracted using a 1:10 dilution of the diarrheal stool sample in SLS. The diluted stool sample was mixed well by votexing for 10 sec and centrifuged for 2 min at 2000xg. One hundred microliters of the supernatant was mixed with 100 μl of lysis solution (15% chelex-100, Brij 58, and 1% Tween 20) and boiled at 95 0C for 10 min. The boiled suspension was vortexed for 10 sec and spun for 2 min at 13,000xg.
DNA Amplification
Two microliters of the clear supernatant was used for the LAMP reaction. This multiplex LAMP method simultaneously amplifies and detects a 240bp region of tcdC gene (this gene is the negative regulator and a surrogate marker for tcdA [toxin A] and/or tcdB [toxin B] genes located in the PaLoc region of the genome), a 222bp region of cdtA (binary toxin gene), and a 223bp region of λ bacteriophage genomic DNA (internal amplification control).
All primers used for amplification and probes used for detection were designed using a public DNA sequence database (Genbank) and PrimerExplorer V4 LAMP primer designing software (Eiken Chemical Co., Ltd. Japan). All synthetic labelled and unlabeled oligonucleotide primers were synthesized by Biosearch Technologies, Inc. (Novato, CA, USA). Oligonucleotide sequences are listed in the supplementary (Table 1).
Table 1 Primers and Probes used for detection of toxigenic C. difficile.
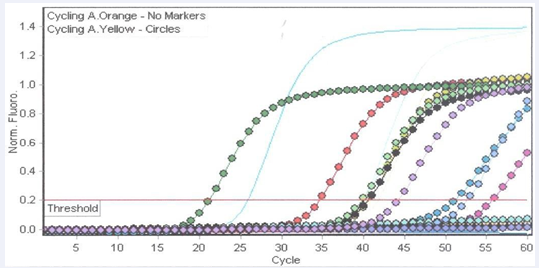
Q-FIP:Fdprimer:probe duplexes were annealed by heating 50 μM Q-FIP and 50 μM Fd to 98 0C and slowly cooling the mixture to room temperature (Figure 1).
Figure 1: Detection of Toxigenic C. difficile using LAMP-DARQ.
LAMP reactions were performed in 1X Isothermal Amplification Buffer (New England Biolabs) : 20mM Tris-HCl (pH8.8), 10mM (NH4)2 SO4 , 50mM KCl, 2mM MgSO4
, 0.1% Tween-20 supplemented with 6 mM MgSO4, and 1.4 mM each of dATP, dCTP, dGTP, and dTTP nucleotides. LAMP reactions contained 0.8 μM FIP, 0.8 μM FIP:Fd, 1.6 μM BIP, 0.2 μM F3 and B3, 0.4 μM LoopF and LoopB, and 0.64U per μl Bst
2.0 WarmStart DNA polymerase (New England BioLabs) [3]. Isothermal DNA amplification and detection was done at 59 0C for 60 min using a Rotorgene 6500 real-time instrument (QIAGEN Inc., Mississauga, ON, Canada). The progress of amplification was detected using reduced quenching (DARQ) as indicated by increased fluorescence from the target probes (Fd); Yellow, oragne and green flourescent signals for tcdC, cdtA and lambda internal control, respectively (Figure 1).
Five microliters of easyMAG™ purified DNA in a 25 μl reaction was used for the PCR. Real-Time PCR was carried out using TaqMan probes as previously reported [4].
RESULTS AND DISCUSSION
Out of 518 specimens tested 200 were positive by PCR and 189 were identified to have toxigenic C. difficile using LAMP. There were 11 discrepant samples and all were negative by LAMP and positive by PCR (Table 2).
Table 2: Comparison of LAMP-DARQ with PCR detection.
Of these 7 had PCR Ct values >37. The other 4 specimens had PCR Ct values between 35 and 37. The test performance characteristics of the LAMP-DARQ method as compared to the PCR are as follows: sensitivity, 95%; specificity, 100%; negative predictive value, 97%; and positive predictive value, 100%. The average turn-around-time for LAMP-DARQ is 1.5 hrs as compared to over 3 hrs for the in-house real-time PCR from the time of specimen reception. The estimated cost per test excluding labor for the LAMP-DARQ method is Cdn $3.75 (nucleic acid extraction, $2.00; amplification and detection, $1.75) and is approximately 60% cheaper than the in-house PCR. The lower limit of detection of the LAMP method was determined by using a ten-fold serial dilution of a known concentration of C. difficile ATCC43255 genomic DNA and was estimated to be 750 genome equivalents per ml of stool. The presence of both tcdC and cdtA presumptively identified 65 specimens to have the NAP1 strain by both methods.
Effect of unpurified sample volume on LAMP detection was examined by adding increasing volumes of control DNA extract to the LAMP reaction. With increasing volume there was an increasing time for detection. However, even with six time volume increase the time to detection was only doubled indicating the robust nature of the Bst I DNA polymerase with minimal inhibition unlike Taq DNA polymerase used for PCR.
Due to the faster turn-around-time, relatively cheaper feature, and ease of operation of LAMP method allow the laboratories to perform testing more frequently at multiple times a day in order to provide results faster to the healthcare facilities to improve not only patient care but also bed management and infection prevention and control especially when there is a high occupancy situation. Often many laboratories employ two step testing algorithms for C. difficile testing. First a screening method using lateral-flow immunoassay to detect C. difficile common antigen and toxin and second a nucleic acid-based method as the confirmatory testing to reduce the operational costs. The desirable fatures described for the LAMP method will allow the laboratories to perform only one test and achieve comparable results faster at a lower operational cost. Since validated the LAMP method described here has been implemented at the Hamilton Regional Laboratory Program for several years performing over 60,000 tests three times a day to improve patient care and to manage and minimize outbreak situations.
CONCLUSION
The method described here is an inexpensive, faster multiplex real-time Loop-Mediated Isothermal Amplification method (LAMP) to detect toxigenic C. difficile and to presumptively identify the hypervirulent NAP1 strain directly from diarrheal stools without nucleic acid purification.