Case Report of Twin Crohn’s Disease Caused by IRF5 Gene Mutation and Literature Review
- 1. Department of Gastroenterology, The Yancheng Clinical College of Xuzhou Medical University, China.
- 2. Faculty of Chinese Medicine, Macau University of Science and Technology, China
- 3. Department of Gastroenterology, Yancheng NO.1 People’s Hospital. China.
Keywords
• Crohn’s Disease
• Inflammatory Bowel Disease
• IRF5/SLEB10
• Interferon Regulatory Factor
• Gene Mutation
Citation
Huang Y, Ye Z, Wang Y, Wu X, Li X (2025) Case Report of Twin Crohn’s Disease Caused by IRF5 Gene Mutation and Literature Review. JSM Gastroenterol Hepatol 12(1): 1133.
Abstract
This paper presents a case of twin brothers afflicted with Crohn’s Disease (CD), characterized by a rare missense mutation c.-12+198G>T in the interferon regulatory factor 5 (IRF5) gene, which is extremely uncommon in the Chinese population and reportedly identified for the first time internationally. The clinical features of the patients are described in detail, and potential pathogenic mechanisms are discussed. In terms of treatment, both twins experienced significant improvement in clinical symptoms and endoscopic findings after treatment with Ustekinumab. Furthermore, through a literature review, this paper anticipates the role of IRF5 in Inflammatory Bowel Disease (IBD) and summarizes its mechanisms, aiming to provide a theoretical basis for future research into personalized treatment of CD.
INTRODUCTION
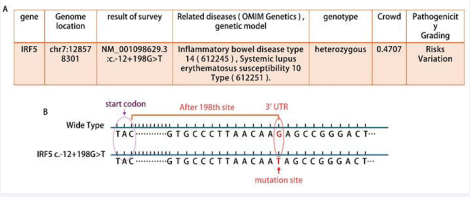
IRF5 belongs to the family of transcription factors, which is initially involved in antiviral response and interferon production [1]. Subsequent studies have used IRF5 as a core regulator of the inflammatory response. A number of studies have revealed the important role of IRF5 in the pathogenesis of a variety of inflammatory and autoimmune diseases, including rheumatoid arthritis (RA), systemic lupus erythematosus (SLE) and IBD [2]. Although the IRF5 gene mutation is rarely reported in IBD patients, a pair of Chinese refractory CD patients (twins) were subjected to whole exome sequencing on July 25,2024. The report showed that they all carried the IRF5 gene c.-12 + 198G > T mutation Figure 1A,B). Sanger sequencing as shown in Figure 1: presents detailed information about a single nucleotide variant (SNV) in the IRF5 gene. “chr7:128578301” indicates that this variant is located at position 128,578,301 on chromosome 7. “NM_001098629.3” is the identifier for the mRNA sequence of this gene. The “c” in “c.-12+198G>T” denotes the cDNA sequence, “-12” signifies that the variant is located 12 base pairs upstream of the start codon, and “+198” indicates that the variant is 198 base pairs downstream of the start codon. At this site, guanine (G) is changed to thymine (T).
Figure 1 Sequencing test results B. IRF5 normal and mutant gene sequences pattern graph
This is a variant within the untranslated region (UTR), which may affect the level of gene expression but does not necessarily alter the amino acid sequence of the protein. Such a variant may exert its effects by influencing mRNA stability, translation efficiency, or interactions with RNAbinding proteins. This mutation has not been reported in domestic or international literature. A summary has been made based on its medication process and symptomatic changes, which also provides valuable references for clinical diagnosis, treatment, and scientific research.
CASE PRESENTATION
Case 1
Male (twins young brother), 26 years old, treated at the Yancheng NO.1 People’s Hospital in 2017 for “perianal mass”. Preliminary colonoscopy suggested CD, but the patient had no symptoms such as abdominal pain or diarrhea. Subsequently, the patient was transferred to the Affiliated Hospital of Nanjing University of Chinese Medicine, where CD was confirmed. The treatment plan included oral “thalidomide” at 50mg, qd. In October 2017, the patient underwent incision and drainage for a perianal abscess at our hospital, with a good postoperative recovery. Thalidomide 50mg qd was maintained for longterm treatment, along with compound glutamic acid enteric-coated capsules and Bifidobacterium quadruple viable tablets to improve intestinal microcirculation. On July 29, 2021, the patient first received vedolizumab (300mg, q8w).
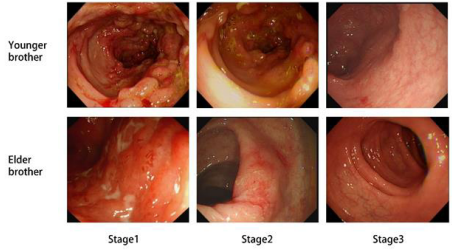
Due to low blood drug concentrations, the dosing interval was adjusted to every 6 weeks, with the last administration on June 7, 2023 (13th dose). In September 2023, the patient was initially treated with ustekinumab (300mg, q8w), completing a total of 4 infusions. Past Medical History: The patient denies a history of hypertension, diabetes, and other conditions. Infectious Disease History: The patient has had hepatitis B for over 4 years and has been taking “Entecavir Dispersible Tablets” 0.5mg qd, and has now transitioned to a Hepatitis B minor three-positive status. The patient denies a history of other infectious diseases such as tuberculosis and malaria. In October 2017, the patient underwent an incision and drainage procedure for a perianal abscess and denies any history of blood transfusion. Colonoscopy Findings: On 2017-10-3 (stage 1), the ileocecal valve was deformed and narrowed, with significant congestion and edema at the terminal ileum. Multiple long and submucosal elevations were visible, with a red surface, and the appendix orifice was absent.
The ascending colon showed uneven-sized cobblestone-like elevations with congested erosions that bled easily upon touch. The rest of the colon had scattered longitudinal shallow depressions, covered with a thin white coating in the center, and the surrounding mucosa was highly congested and edematous, with scattered multiple ulcers and erosions of varying sizes.
The rectum showed scattered erosions and bleeding points. On 2022-8-18 (stage2),the ileocecal valve was deformed, with congestion in the terminal ileum mucosa. The ascending colon had multiple uneven-sized cobblestone-like elevations, pseudopolyp formation, and scattered erosions with longitudinal white scars visible. On 2024-6-11 (stage3), the ileocecal valve was deformed, with edema in the terminal ileum mucosa.
Figure 2 Colonoscopy findings of two twin brothers at different stages of medication
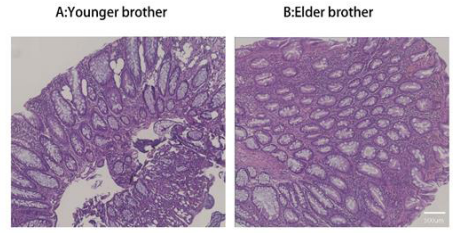
The ascending colon showed a few scattered flat congested patches and longitudinal white scars, with no significant abnormalities observed in the rest of the colon (Figure 2). Pathology Examination (stage2): Chronic active enteritis with local mucosal erosions, a small number of epithelioid granulomas, and local lymphoid follicle formation (Figure 3A).
Case 2
Male (twins elder brother), 26 years old, the patient visited the Yancheng NO.1 People’s Hospital on October 5, 2019, due to “bloody mucus in the stool” and was diagnosed with “Crohn’s disease,” with recurrent symptoms of bloody stools. There was no abdominal pain or diarrhea. On July 27, 2021, he received his first treatment with vedolizumab (300mg, q8w) and has been taking the medication regularly, with the last use of vedolizumab on May 15, 2023. On September 1, 2023, he first used ustekinumab (300mg, q8w) and has used it three times. Past Medical History: Denies history of hypertension, diabetes, and other medical conditions.
Has a history of hepatitis B, denies history of typhoid, tuberculosis, and other infectious diseases. Had a perianal abscess surgery in 2016, from which he has recovered. Denies history of blood transfusion. Colonoscopy Findings: On July 23, 2021 (stage1), scattered erythematous erosions were observed in the terminal ileum mucosa, and multiple inflammatory polyps with irregular shallow depressions were seen throughout the colon, with a thin white coating at the center and congested edema at the edges, some showing longitudinal ulcers. The rectal mucosa was focally rough and congested with clear vascular patterns, with no obvious ulcers or neoplasms.
On June 22, 2023 (stage2), scattered small patches of erosions and shallow ulcers were seen in the terminal ileum. The entire colon showed multiple flat erythematous erosions and white longitudinal scar formations. No significant abnormalities were observed in the rectum. On June 5, 2024 (stage3), a few scattered longitudinal white scars were seen throughout the colon, with no significant abnormalities observed in the rest of the terminal ileum and colon (Figure 2). Pathological Examination (stage2): Chronic active enteritis with an increase in chronic inflammatory cells in the mucosal propria, and local lymphoid follicle formation (Figure 3B).
Figure 3 (Stage2): Pathological examination: hematoxylin-eosin stages of intestinal mucosa in twin brothers.
LITERATURE REVIEW
Searches were conducted in PubMed, CNKI (China National Knowledge Infrastructure), and Wanfang databases using the keywords “Crohn’s disease,” “IRF5,” “inflammatory bowel disease,” As of September 1, 2024, after screening and excluding duplicates, reviews, and literature not in Chinese or English, a total of three relevant articles were obtained.
A meta-analysis delved into the close relationship between the IRF5 rs4728142 genotype and IBD, revealing the potential role of this genotype in the pathogenesis of IBD [3]. Another study focused on two groups of IBD patients from the North American consortium, systematically investigating six single nucleotide polymorphisms (SNPs) and five base pair (bp) insertion-deletion (CGGGG indel), polymorphisms. The study found that specific dual-marker haplotypes (rs4728142-CGGGG indel), and (CGGGG indelrs7808907), are associated with protective effects and risks for IBD, providing a new perspective for genetic research in IBD [4]. The third article genotyped 12 polymorphisms of the IRF5 gene in a cohort of 1007 IBD patients and 241 controls from the Walloon region of Belgium, as well as in 311 controls and 687 IBD patients from Leuven, Belgium. A strong association signal was observed for the 5bp insertion-deletion (CGGGG), polymorphism in the promoter region of the IRF5 gene with IBD, indicating that the insertion-deletion polymorphism of the interferon regulatory factor 5 (IRF5) gene is closely related to the risk of developing IBD [5]. These studies not only enrich our understanding of the genetic background of IBD but also provide a scientific basis for future precision medicine and personalized treatment.
DISCUSSION
Interferon regulatory factors (IRFs) are a family of transcription factors that regulate immune responses, initially discovered for their role in mediating antiviral responses and the production of type I interferons (IFNs). Subsequently, it has been confirmed that they also play various functions in apoptosis, cell cycle, tumor development, and gene regulation in response to pathogen signals [1]. The IRF family includes nine members (IRFs1- 9), which have a conserved multi-domain structure [6]. The N-terminal DNA-binding domain (DBD) recognizes the core DNA sequence in the interferon-stimulated response elements, and the C-terminal IRF association domain (IAD) mediates protein-protein interactions between IRFs and other proteins, forming transcription complexes [7,8]. The IRF5 protein is a 60-63kDa polypeptide with a typical IRF structure, consisting of a DBD and an IAD. The DBD is characterized by the presence of five tryptophan residues, which fold into a winged helix-turn-helix conformation, a structure that is crucial for its DNA-binding activity [9].
Among the members of the IRF family, IRF5 plays a central role in inflammation. IRF5 activates three major inflammatory signaling pathways, including NF-κB, MAPK, and IRF, which are key in inflammation. IRF5 not only regulates the type I IFN system but also induces the expression of various pro-inflammatory cytokines, such as IL-6, IL-12, IL-23, and TNF-α, which are also crucial in IBD. Furthermore, IRF5 assists the p65 subunit (RelA) of the NF-kB complex in recruiting to the promoter regions of inflammatory genes, an interaction that helps to enhance the expression of these genes [10,11]. IRF5 is also a key factor in determining the phenotype of inflammatory macrophages. IRF5 can induce the polarization of macrophages towards the M1 phenotype, potentially exacerbating symptoms in IBD patients with comorbid depression.
Experiments have shown that in DSS-induced colitis and depression in mice, the expression of IRF5, CD86, and inflammatory factors in the intestinal mucosa is elevated compared to non-depressed mice. Knocking down IRF5 can shift the polarization of rat peritoneal macrophages from the M1 phenotype to the M2 phenotype, suppressing inflammation [12]. It is highly expressed in monocytes, macrophages, B cells, and dendritic cells, particularly when stimulated by GM-CSF and IFN-γ, leading to a significant upregulation of IRF5 in macrophages and thus enhancing the inflammatory response [11,13]. IRF5 has garnered significant attention due to its role in regulating inflammatory responses and autoimmune diseases. It can modulate immune cell functions, and its dysregulation may lead to immune disorders. Although IRF5 has been largely implicated in the pathogenesis of SLE and RA [14-16], its connection with IBD is also gaining attention. Studies have shown that IRF5 is overexpressed in the intestinal mucosa of IBD patients, with its expression levels positively correlating with IBD disease activity, and it may serve as a potential biomarker for IBD management by regulating Th1 and Th17 cell immune responses and cytokine production [17]. Multiple studies have revealed the association between IRF5 gene polymorphisms and the risk of IBD in different ethnicities. A deeper understanding of how different IRF5 gene polymorphisms exert protective or detrimental effects on IBD patients of various ethnicities is not only significant for assessing the risk of IBD but may also provide new perspectives and methods for IBD treatment strategies.
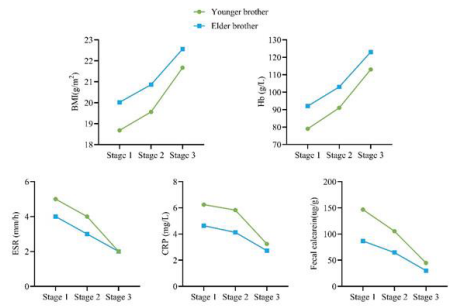
The IRF5 gene mutation in this pair of twins is reported for the first time in medical literature. By comparing the disease characteristics of the two brothers, we can identify the following similarities and differences. Similarities: 1. Both carry the same IRF5 gene mutation: NM_001098629.3:c.-12+198G>T. 2. According to the Montreal classification, both are diagnosed with A2L3B1p type. 3. After treatment with ustekinumab, both showed Significant improvement in their conditions (Figure 4 for detailed treatment changes). 4. Both have suffered from perianal abscesses, a common complication of CD. 1. Differences: There is a significant difference in the severity of their conditions: the mean CDAI score for the twin brother is 380, while for the twin brother, it is 160. Additionally, CRP, ESR, fecal calprotectin, and BMI differ, as shown in Figure 4. 2. Their age of onset is different. These findings not only provide a new perspective on the role of IRF5 gene mutations in Crohn’s disease but also emphasize that even individuals with the same genes can exhibit differences in disease manifestation and treatment response. This information is of significant importance for the development of personalized medicine and precision treatment strategies.
Figure 4 Treatment process: stage 1: Initial onest stage 2: after adalimumab treatment. Stage 3: after treatment with unlinastatinumab.
integrin α4β7, which specifically binds to α4β7 integrins at intestinal inflammatory sites, blocking their interaction with the adhesion molecule MAdCAM-1, reducing the migration of inflammatory cells to the inflamed intestinal mucosa, and achieving an anti-inflammatory effect. Vedolizumab is suitable for CD and ulcerative colitis (UC) patients who are unresponsive to conventional treatments. However, due to the IRF5 gene mutation carried by these twin patients, this mutation may lead to changes in protein function. Studies have shown that IRF5 can activate multiple downstream inflammatory pathways such as NF-κB, MAPK, and IRF, as well as promote the expression of pro-inflammatory cytokines IL-6, IL-12, and TNF-α. Therefore, we reasonably speculate that vedolizumab may not affect the pathogenesis of CD in these twins, which could also be the reason they did not achieve the desired effect with previous treatments.
Ustekinumab is a fully human IgG1κ monoclonal antibody primarily acting as an IL-12/IL-23 p40 inhibitor for the treatment of plaque psoriasis and CD. It specifically binds with high affinity to the p40 protein subunit of IL-12 and IL-23, blocking the interaction of these cytokines with the cell surface receptor IL-12Rβ1, thereby inhibiting IL12 and IL-23-mediated signal transduction and cytokine cascade reactions, achieving targeted immunological antiinflammatory effects. Given that IRF5 can activate the proinflammatory cytokine IL-12, the significant improvement in the patients’ clinical symptoms and endoscopic findings after switching the treatment regimen from vedolizumab to ustekinumab is observed.
Figure 5 The changes of BMI, Hb, ESR, CRP, and fecal calprotectin in twins at different stages of medication.
Based on these observations, subsequent treatments could consider pharmacological therapies targeting inflammation pathways affected by IRF5 for better therapeutic outcomes. Such personalized treatment plans may provide more effective treatment options for Crohn’s disease patients, especially those with IRF5 gene mutations .
Considering the crucial role of the IRF5 gene in IBD, it is particularly important to delve into the impact of the specific point mutation c.-12+198G>T in the pathogenesis and progression of IBD. Through meticulous scientific research, we can reveal how this mutation affects the function of the IRF5 protein and its involvement in regulating inflammatory responses. This will not only help us to understand the complex pathological processes of IBD more comprehensively but also provide patients with more personalized and targeted treatment plans. The development of specific drugs or treatment strategies targeting IRF5 mutations may also bring new hope for IBD patients. By precisely targeting inflammation pathways related to IRF5, we can more effectively control the condition, reduce side effects, and improve treatment outcomes.
This molecular-level personalized medical strategy is expected to provide IBD patients with more precise and efficient treatment options. In exploring treatment options, we can consider innovative therapeutic methods, including hematopoietic stem cell transplantation and fecal microbiota transplantation. Hematopoietic stem cell transplantation, as a potential treatment, may suppress abnormal inflammatory responses by resetting the immune system. Fecal microbiota transplantation, by introducing a healthy gut microbiome, may regulate the patient’s intestinal environment and thus may have a positive impact on the treatment of IBD.
REFERENCES
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008; 26: 535-584.
- Almuttaqi H, Udalova IA. Advances and challenges in targeting IRF5, akey regulator of inflammation. Febs J 2019; 286: 1624-1637.
- Tang L, Xu M. Candidate polymorphisms and susceptibility to inflammatory bowel disease: A systematic review and meta-analysis. Gene. 2020; 753: 144814.
- Gathungu G, Zhang CK, Zhang W, Cho JH. A two-marker haplotype in the IRF5 gene is associated with inflammatory bowel disease in a North American cohort. Genes Immun. 2012; 13: 351-355.
- Dideberg V, Kristjansdottir G, Milani L, Libioulle C, Sigurdsson S, Louis E, et al. An insertion-deletion polymorphism in the interferon regulatory Factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Hum Mol Genet. 2007; 16: 3008-3016.
- Mamane Y, Heylbroeck C, Génin P, Algarté M, Servant MJ, LePage Ceta. Interferon regulatory factors: the next generation. Gene. 1999; 237: 1-14.
- Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, HakoshimaT. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. Embo J. 1999; 18: 5028-5041.
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001; 19: 623-655.
- Zhu KC, Guo HY, Zhang N, Liu BS, Guo L, Jiang SG, et al. Structural and expression analysis of golden pompano Trachinotus ovatus IRF5 and its role in regulation of type I IFN. Fish Shellfish Immunol. 2020; 97: 313-321.
- Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005; 434: 243-249.
- Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011; 12: 231-238.
- Liang C, Tang Y, Gao X, Lei N, Luo Y, Chen P, et al. Depression Exacerbates Dextran Sulfate Sodium-Induced Colitis via IRF5- Mediated Macrophage Polarization. Dig Dis Sci. 2023; 68: 1269-1279.
- Weiss M, Blazek K, Byrne AJ, Perocheau DP, Udalova IA. IRF5 is a specific marker of inflammatory macrophages in vivo. Mediators Inflamm. 2013; 2013: 245804.
- Ban T, Kikuchi M, Sato GR, Manabe A, Tagata N, Harita K, et al. Genetic and chemical inhibition of IRF5 suppresses pre-existing mouse lupus-like disease. Nat Commun. 2021; 12: 4379.
- Song S, De S, Nelson V, Chopra S, LaPan M, Kampta K, et al. Inhibition of IRF5 hyperactivation protects from lupus onset and severity. J Clin Invest. 2020; 130: 6700-6717.
- Cie?la M, Kolarz B, Majdan M, Darmochwa?-Kolarz D. IRF5 promoter methylation as a new potential marker of rheumatoid arthritis. Pol Arch Intern Med. 2019; 129: 370-376.
- Yang Y, Zhang C, Jing D, He H, Li X, Wang Y, et al. IRF5 Acts as a Potential Therapeutic Marker in Inflammatory Bowel Diseases. Inflamm Bowel Dis. 2021; 27: 407-417.