MRPL3 Promotes Gastric Cancer Progression through Inhibiting the Ubiquitination of EGFR
- 1. The First Hospital of Ningbo University, China
- 2. The Second Affiliated Hospital of Harbin Medical University, China
- 3. Heilongjiang Provincial Second Tumor Hospital, China
Abstract
Background: As a common and lethal malignancy, gastric cancer remains in urgent need of investigation of its molecular mechanisms and effective molecular targets for therapy to improve patient outcomes. MRPL3, a mitochondrial ribosomal protein, plays a critical role in protein synthesis within the mitochondrion. A number of studies have reported that MRPL3 is involved in various fatal diseases, including cancers. However, the role of MRPL3 in gastric cancer remains largely elusive.
Methods: Here, we assigned 249 tissues from patients who had undergone surgical resection of early gastric cancer at The Affiliated Hospital of Medical School, Ningbo University. We examined the expression and prognostic value of MRPL3 in gastric cancer patients by using tissue microarrays, immunohistochemistry, and survival analysis. We also investigated the molecular effect of MRPL3 on gastric cancer via RNA-seq, RT-qPCR, Western blot, coimmunoprecipitation, and ubiquitination assays. Furthermore, we explored the interaction between MRPL3 and EGFR, a key factor in gastric cancer progression, and elucidated its impact on EGFR signaling and degradation.
Results: We found that MRPL3 was overexpressed and correlated with poor survival in gastric cancer and that MRPL3 knockdown significantly impeded the proliferation, invasion, and tumor growth of gastric cancer. Furthermore, MRPL3 activated the EGFR/STAT1 signaling pathway by inhibiting the ubiquitination and degradation of EGFR, and knockdown of MRPL3 had an inhibitory effect on the progression of gastric cancer. Finally, the upregulation of EGFR expression reversed the anti-gastric cancer effect of MRPL3 knockdown both in vitro and in vivo.
Conclusion: Our study reveals a novel mechanism by which MRPL3 plays an indispensable role in promoting gastric cancer progression by activating the EGFR/STAT1 signaling pathway through inhibiting the ubiquitination and degradation of EGFR. Therefore, MRPL3 could be a potential therapeutic target for gastric cancer.
Citation
Chena R, Jianga H, Qiu S, Quan S, Luo K, et al. (2025) MRPL3 Promotes Gastric Cancer Progression through Inhibiting the Ubiquitination of EGFR. JSM Gastroenterol Hepatol 12(1): 1134.
BACKGROUND
Gastric cancer is the third most common leading cause of cancer mortality globally [1,2]. Endoscopic biopsy, CT, PET, laparoscopy and endoscopic ultrasound are used to histologically diagnose and stage gastric cancer [1]. Endoscopic resection remains the only treatment that offers curative potential for early gastric cancer, whereas gastric cancer generally carries a poor prognosis because most gastric cancer patients often lose the chance of treatment for diagnosis at an advanced stage [3]. Moreover, gastric cancer at an advanced stage is commonly treated with sequential lines of systemic therapy, which can improve survival and enhance the quality of life of patients with locally advanced or metastatic disease [4].
Currently, molecular targeted therapy shows great potential to improve cancer patient survival but lacks molecular targets [4,5]. Thus, there is an urgent need to develop potential therapeutic targets and therapeutic strategies for the treatment of gastric cancer. Mitochondrial ribosomal protein L3 (MRPL3), also known as MRL3, RPML3 or COXPD9, belongs to the L3P ribosomal protein family and consists of a 39S subunit protein, which is implicated in protein synthesis within the mitochondrion [6]. It has been reported that MRPL3 is involved in various fatal diseases that threaten human health, including adult-onset neurodegeneration [7], neurological disorders [8], mitochondrial cardiomyopathy [9] and cancers (breast cancer [10], colorectal cancer [11], and lung cancer [12,13]). For instance, MRPL3 was identified as a novel hub gene of breast cancer through WGCNA and a literature search, which means that MRPL3 can be a promising candidate gene and serve as a diagnostic and prognostic biomarker of breast cancer [10].
These works revealed MRPL3 as an aberrant molecule in the maintenance of the development of some malignant cancers, whereas the exact biological role of MRPL3 in gastric cancer and the relevant mechanism deserve further in-depth study. Epidermal growth factor receptor (EGFR), as an activator of STAT1, has been reported to be aberrantly overexpressed and to promote tumorigenesis of gastric cancer [14-16]. A number of works have shown that aberrant activation of EGFR plays a critical role in the tumorigenesis and invasion of gastric cancer by modulating the EGFR/STAT1 signaling pathway [17,18].
However, the upstream regulators that participate in the overactivated EGFR/STAT1 signaling pathway in gastric cancer remain elusive (Figure 1). In our study, we uncovered that overexpressed MRPL3 was correlated with poor survival in gastric cancer patients. Furthermore, silenced MRPL3 could decrease the proliferative and invasive abilities of gastric cancer cells significantly by enhancing the degradation of EGFR, which was proven to rely on the ubiquitination regulated by MRPL3. Taken together, our results emphasize the importance of MRPL3 as a therapeutic target in gastric cancer.
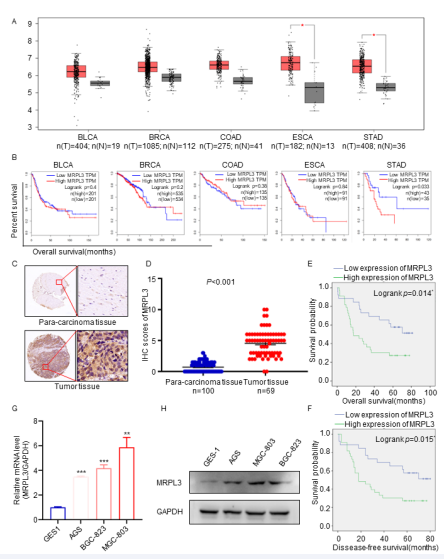
Figure 1 Overexpression of MRPL3 is correlated with an unfavorable prognosis in gastric cancer. A. The GEPIA database was searched for MRPL3 mRNA expression in several malignant cancers. Bladder urothelial carcinoma (BLCA), Breast invasive carcinoma (BRCA), Colon adenocarcinoma (COAD), Esophageal carcinoma (ESCA), Stomach adenocarcinoma (STAD). * , p<0.05. B. The overall survival data for several malignant cancer patients with high or low MRPL3 expression levels in the GEPIA database. P values are shown in Fig. C. IHC images of MRPL3 staining in TMA tissue sections are shown. The scale bars are shown in panel D. Dot plots show the IHC scores of MRPL3 expression for TMA tissue sections (normal gastric specimens: n=100, gastric TMA specimens: n=69, p<0.001). Statistical analyses were performed with the D’Agostino & Pearson omnibus normality test. E-F. According to Kaplan?Meier survival analysis, overexpressed MRPL3 was significantly correlated with disease-free survival and overall survival of gastric cancer patients. G and H. RNA and protein expression of MRPL3 in normal gastric cells and gastric cancer cells.
MATERIALS AND METHODS
Cell culture
The AGS, MCG-803, GES1 and BGC-823 cell lines were purchased from the Type Culture Collection Cell Bank of the Chinese Academy of Sciences (Shanghai, China). GES1 and MGC-803 cells were cultured in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA)) and 1% penicillin/streptomycin at 37 °C in a 5% CO2 incubator. AGS and BGC-823 cells were cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA)) and 1% penicillin/ streptomycin at 37 °C in a 5% CO2 incubator.
Antibodies and Chemical Reagents
Total cell lysates were obtained from the indicated cells with RIPA lysis buffer containing 1% protease and phosphatase inhibitors (Sigma?Aldrich) on ice for >30 min. The resulting cell lysates were centrifuged for 15 min at 13,000 rpm at 4 ? to remove undissolved impurities, and whole protein supernatants were collected. The concentration of total cell protein was quantified using a BCA assay (#P0012S, Beyotime). Protein extracts were incubated with appropriate primary antibody beads overnight for an immunoprecipitation assay or evaluated for western blot analysis. The precipitated immune complexes were separated by 10%-15% sodium dodecyl sulfate?polyacrylamide gel electrophoresis (SDS?PAGE), transferred to 0.45-μm polyvinylidene difluoride (PVDF) membranes, blocked with 5% silk milk at room temperature for 1 h and incubated with specific primary antibodies at 4 °C overnight. Autoradiograms were quantified by ImageJ software.
Cell Viability
Assay Cell proliferation was assessed by a CKK8 assay (Beyotime, China) at the indicated times. The OD450 was determined by a microplate reader (REAGEN, RNE90002, USA).
Colony Formation Assay
Cells (800 cells/well) were seeded in a 12-well cell plate and cultured for 2 weeks. Then, the cells were fixed with 4% paraformaldehyde Fix Solution for 30 min at room temperature after washing the plate 3 times. Then, the plate was lightly washed 3 times with PBS, stained with crystal violet, observed and photographed under an optical microscope. The number of colonies was counted by ImageJ software. All assays were performed in triplicate.
Migration Assay
shCtrl-AGS, shMRPL3-AGS, shCtrl-MGC803 and shMRPL3-MGC803 cells were cultured in upper chambers (8-μm pore size; 3422 Corning) with serum-free medium, and 20% FBS medium was placed in the lower chambers. After culturing for 12-24 hours, the cells on the bottom surface of the membrane were fixed with 4% paraformaldehyde Fix Solution for 30 mins at room temperature and stained with crystal violet solution for 30 mins. The migrated cells were photographed under a microscope and counted using ImageJ software.
Quantitative RT?PCR Assay
Total RNA was extracted with TRIzol. RNA reverse transcription was performed with the PrimeScript RT Reagent Kit (#RR047A, TAKARA, Japan). Real-time PCR was conducted with a qPCR SYBR Green Master Mix (11198ES03, YEASEN, China). The 2-ΔCt method was used to quantify fold changes with normalization to GAPDH. Detailed information on the primer sequences is shown in [Table S1].
Table S1: Antibodies and Chemical Reagents.
|
Antibody name |
Working dilution |
Item No. |
Purchased from |
|
TRIM2 |
1:500 |
SRP09319 |
Tianjin Saierbio |
|
EGFR |
1:4000 |
66455-1-Ig |
Proteintech |
|
STAT1 |
1:1000 |
10144-2-AP |
Proteintech |
|
SCL7A11 |
1:500 |
bs-6883R |
bioss |
|
PRKACB |
1:10000 |
ab76238 |
Abcam |
|
GAPDH |
1:30000 |
60004-1-lg |
Proteintech |
|
Goat Anti-Rabbit |
1:3000 |
A0208 |
Beyotime |
|
Goat Anti-Mouse |
1:3000 |
A0216 |
Beyotime |
RNA Interference
The shRNAs of MRPL3 were purchased from Gene Pharma. The sequences were as follows:
shMRPL3-1, 5’- GACTGAAAGTGTGGAGAATAA -3’;
shMRPL3-2, 5’- GCATATAAGGATCTCGGTAAA -3’;
shMRPL3-3, 5’- GTCTTCATGGAAAGAGTGGTA -3’.
Generation of the Tumor Xenograft Model
BALB/c nude mice (six weeks old) were purchased from HFK Bioscience and raised under SPF conditions. For the generation of the subcutaneous xenograft model, shCtrlAGS and shMRPL3-AGS cells (1×106/mouse) were injected subcutaneously into mice. The mouse weights and tumor volumes were detected every day and calculated by using the following formula: (L × W2/2). Mice were euthanized after 12 days, and xenograft tumors were harvested. All animal studies in the present work were approved by the related committee of the Affiliated Hospital of Medical School, Ningbo University.
Immunohistochemistry (IHC)
IHC staining was performed as previously described [19]. The staining intensity was divided into four grades (A: 0, negative; 1, weakly positive; 2, positive; 3, strongly positive), and the percentage of staining-positive cells was indicated by five grades (B: 0, 75%). The final expression scores were calculated by multiplying A by B.
Statistical Analysis
Statistical analysis was performed by using GraphPad Prism Software. One-way ANOVA or two-tailed Student’s t test was used to analyze the significant differences. All data are presented as the means ± SDs, and P < 0.05 was defined as statistically significant.
RESULTS
Overexpressed MRPL3 was correlated with poor survival in Gastric Cancer Patients
cancers, the TCGA database was searched, and the results indicated that MRPL3 was overexpressed significantly in several malignant cancers, including gastric cancer [Fig 1A]. Moreover, the survival rate analysis results showed that the elevated expression of MRPL3 was correlated with poor overall survival in gastric cancer but not in other malignant cancers [Fig 1B]. Therefore, we speculated that MRPL3 plays a critical biological role in the progression of gastric cancer. Additionally, IHC analysis verified that MRPL3 was overexpressed in gastric cancer patients [Fig 1C-D], and according to Kaplan?Meier survival analysis, MRPL3 gene expression was significantly correlated with overall survival and disease-free survival of gastric cancer; that is, with the increase in MRPL3 gene expression, survival decreased [Fig 1E-F]. Finally, compared with the normal gastric epithelial cell line GES1, MRPL3 in gastric cancer cells, including AGS, BGC823 and MGC803 cells, showed high RNA and protein expression levels [Fig 1G-H]. Therefore, we concluded that MRPL3 was overexpressed significantly and correlated positively with poor survival in gastric cancer.
Silencing MRPL3 inhibited the Proliferation and Invasion of Gastric Cancer Cells in Vitro
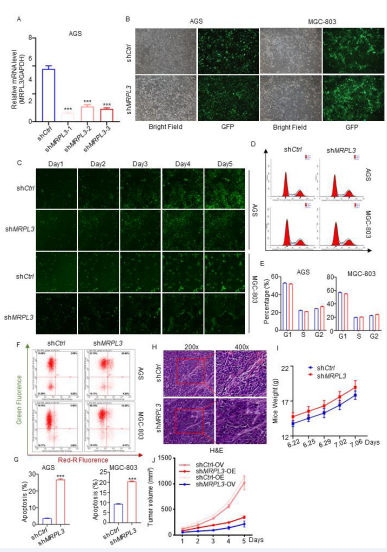
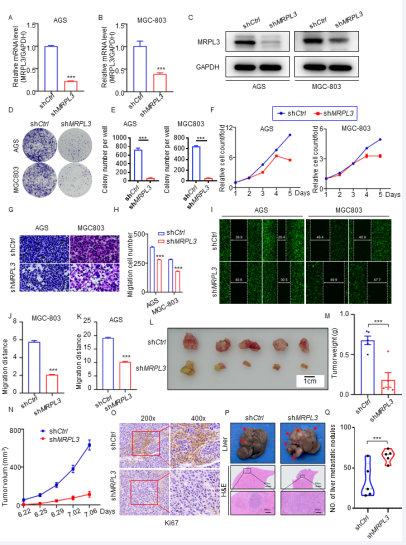
To explore the biological role of MRPL3 in the tumorigenesis of gastric cancer, we established gastric cancer cell lines with silenced MRPL3 [Fig 2A-C, S2A-B]. Colony formation assays showed that MRPL3 inhibition significantly inhibited the proliferation of gastric cancer cells [Fig 2D-E], and Celigo cell count assays proved that compared with the shCtrl group, the experimental group showed a significant reduction in cell proliferation over the course of the assay [Fig 2F, S2C]. In addition, the results of flow cytometry detection of the cell cycle showed that the percentage of shMRPL3 cells decreased in S phase (P<0.01) and increased in G2 phase (P<0.01) compared with the shCtrl group [Fig S2D-E]. Additionally, a cell apoptosis detection assay proved that compared with the shCtrl group, the AGS and MCG-803 cell apoptosis percentages were significantly increased in the shMRPL3 group [Fig S2F-G].
Figure S2 A. RT-PCR analyses of MRPL3 expression in AGS cells infected with shCtrl, shMRPL3-1, shMRPL3-2, and shMRPL3-3. B. The cell fluorescence of AGS and MGC-803 cells infected with shCtrl and shMRPL3-1. C. The cell fluorescence of AGS and MGC-803 cells infected with shCtrl and shMRPL3-1 and cultured for 5 days under Celigo cell count assays. D-E. Flow cytometry of detection and analyses of Cell Cycle in AGS and MGC-803 cells infected with shCtrl, shMRPL3-1. F-G. Cell apoptosis detection and analyses of of Cell Cycle in AGS and MGC-803 cells infected with shCtrl, shMRPL3-1. H-I. The H&E of tumor and weight of mice. Statistical analyses were performed with one-way ANOVA followed by Tukey’s mulple comparison’s tests. *, p<0.1; **, p<0.01; ***p<0.001.
Moreover, transwell assays and woundhealing proved that MRPL3 inhibition significantly decreased the invasive ability of gastric cancer cells [Fig 2G-J]. To further investigate the biological role of MRPL3 in gastric cancer in vivo, AGS-shCtrl and AGS-shMRPL3 cells were subcutaneously injected into BALB/c nude mice using a xenograft assay under the same conditions. As expected, the tumors formed by AGS-shMRPL3 cells were significantly slower and lighter than those formed by AGS-shCtrl cells [Fig 2L-N, S2H-J]. Immunohistochemical analysis of Ki67 revealed that the underlying mechanism of this phenomenon is that inhibition of MRPL3 significantly suppresses tumor proliferation in vivo [Fig 2 O]. To further examine the effect of MRPL3 on the migration ability of gastric cancer in vivo, we subcutaneously injected AGSluciferase-shCtrl and AGS-luciferase-shMRPL3 cells into the spleens of nude mice to establish a liver metastasis model of gastric cancer. As [Fig 2P-O] shows, the nodes formed in the spleen by AGS-shMRPL3 cells were smaller and fewer than those formed by AGS-shCtrl cells. Taken together, our results suggested that MRPL3 knockdown significantly inhibited the proliferation and invasion ability of gastric cancer cells in vitro and in vivo.
Figure 2 Silencing MRPL3 suppresses the proliferation and aggressive behavior of gastric cancer cells in vitro and in vivo.
Silencing MRPL3 decreases EGFR expression in Gastric Cancer Cells
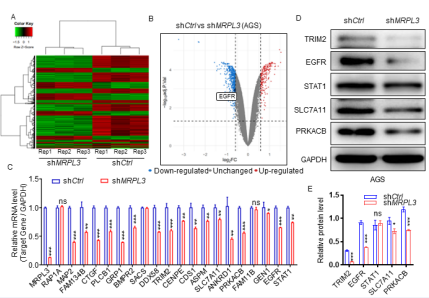
We have proven that silenced MRPL3 could impede the proliferation and invasion of gastric cancer in vitro and in vivo, and it was significantly correlated with poor survival in gastric cancer patients. However, the specific regulatory mechanism underlying these effects is still unclear. By silencing the expression of MRPL3 in AGS cells and performing an RNA-seq assay, we identified 1701 differentially expressed genes (DEGs), including 557 upregulated DEGs and 1144 downregulated DEGs [Fig 3A-B]. Furthermore, we found that EGFR was significantly downregulated after MRPL3 silencing [Fig 3B], which indicated that EGFR could be regulated by MRPL3. Consistently, RT?PCR and western blot analyses indicated that both the mRNA and protein levels of EGFR and some other genes (selected from the results of RNAseq randomly) were significantly downregulated in MRPL3-silenced gastric cancer cells [Fig 3C-E]. These results indicated that silencing MRPL3 could decrease the expression of EGFR in gastric cancer cells.
Figure 3 MRPL3 transcriptionally increases EGFR expression in gastric cancer cells.
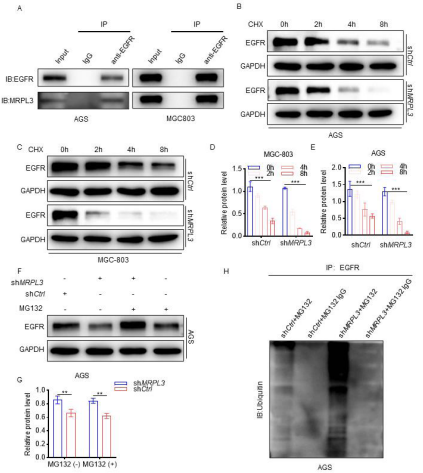
MRPL3 inhibits EGFR Ubiquitination and Degradation
To investigate how MRPL3 regulates the EGFR signaling pathway, we conducted a coimmunoprecipitation (CoIP) assay to investigate whether MRPL3 interacts with EGFR. The results showed that MRPL3 was efficiently precipitated by an antibody against EGFR in AGS and MGC803 cell lines [Fig 4A]. Furthermore, we treated AGS-shMRPL3 and MGC803-shMRPL3 cells with CHX, and the experimental results revealed that compared with shCtrl, downregulation of MRPL3 facilitated the degradation of EGFR in AGS and MGC-803 cells [Fig 4B-E]. Interestingly, compared with AGS-shMRPL3 cells without MG132 treatment, MG132-treated AGS-shMRPL3 cells exhibited increased expression of EGFR [Fig 4F-G]. We noticed that MRPL3 might regulate EGFR ubiquitination and degradation in gastric cancer cells. To validate this hypothesis, we next quantified the relative ubiquitin-EGFR(Ub-EGFR) level by immunoprecipitation with an antiEGFR antibody under MG132 treatment, and the results implied that the ubiquitination of EGFR was enhanced significantly in AGS-shMRPL3 cells [Fig 4H]. These data are consistent with the notion that MRPL3 regulates the ubiquitination and degradation of EGFR in gastric cancer.
Figure 4 MRPL3 inhibited EGFR ubiquitination and degradation.
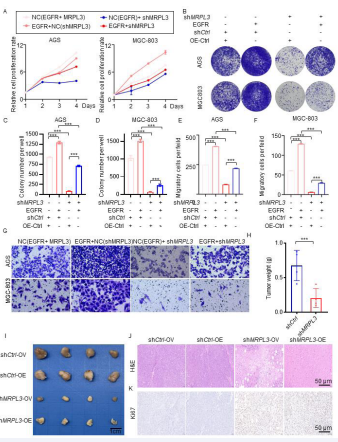
MRPL3 knockdown enhanced the anti-gastric cancer effect via EGFR inhibition in Vivo and in Vitro
To examine the biological role of the MRPL3-EGFR pathway in the tumorigenesis of gastric cancer cells, we established cell models including AGS (shCtrl-OV, shCtrlOE, shMRPL3-OV, shMRPL3-OE) and MGC803 (shCtrl-OV, shCtrl-OE, shMRPL3-OV, shMRPL3-OE) cells. The CCK8 assay indicated that overexpressed EGFR could reverse the inhibited proliferation of gastric cancer cells under MRPL3 inhibition [Fig 5A], and the colony formation assay proved this notion [Fig 5B-C]. Furthermore, the results of the transwell assay implied that overexpression of EGFR could significantly reverse the decreased invasive ability of gastric cells with MRPL3 silencing [Fig E-G]. To further explore the biological role of the MRPL3-EGFR pathway in gastric cancer in vivo, AGS (shCtrl-OV, shCtrl-OE, shMRPL3- OV, shMRPL3-OE) cells were subcutaneously injected into BALB/c nude mice via a xenograft assay under the same conditions. As shown in [Fig 5H-L], the tumors formed by AGS-EGFR-shMRPL3 cells were significantly faster and heavier than those formed by AGS-OV-shMRPL3 cells, which means that EGFR could reverse the anti-gastric cancer effect of MRPL3 inhibition in vivo. These data are consistent with the notion that the enhanced anti-gastric cancer effect in vivo and in vitro via knockdown of the expression of MRPL3 could occur through EGFR inhibition.
Figure 5 MRPL3 induced the proliferation and aggressive behavior of gastric cancer by inhibiting the ubiquitination and degradation of EGFR. AGS and MGC803 cells were infected with shCtrl+OV, shCtrl+OE, shMRPL3+OV, and shMRPL3+OE. The analyses for the CKK8 assay (A) and harvested for colony formation (B-D) and transwell invasion assays after 48 h of culture (E-G). Statistical analyses were performed with one-way ANOVA followed by Tukey’s multiple comparison tests. *, p<0.1; **, p<0.01; ***p<0.001. H-L. AGS (?shCtrl-OV, shCtrl-OE, shMRPL3-OV, shMRPL3-OE) cells were subcutaneously injected into BALB/c nude mice in a xenograft assay. The tumors were harvested and photographed (I) on day 21. Data for tumor weight (H), H&E (J) and IHC for ki67 of tumors (K) are shown as the mean + SEM (n=4). Statistical analyses were performed with one-way ANOVA followed by Tukey’s multiple comparison tests. *, p<0.1; **, p<0.01; ***p<0.001.
DISCUSSION
In this investigation, we explored the pivotal role of mitochondrial ribosomal protein L3 (MRPL3) in gastric cancer progression and its interplay with the epidermal growth factor receptor (EGFR) pathway. Through an extensive array of in vitro and in vivo assays, we observed a marked elevation of MRPL3 expression in gastric cancer specimens, correlating significantly with adverse patient utility of MRPL3 as a prognostic biomarker for gastric cancer. Our findings reveal a previously unidentified mechanism by which MRPL3 fosters gastric cancer cell proliferation and invasion via modulation of the EGFR/ STAT1 signaling axis. Specifically, our data show that MRPL3 impedes the ubiquitination and subsequent degradation of EGFR, thereby amplifying EGFR-mediated signaling and promoting oncogenic growth and invasiveness.
This insight offers a fresh perspective on the molecular drivers of gastric cancer progression and earmarks MRPL3 as a viable target for therapeutic intervention. Considering the instrumental role of MRPL3 in the advancement of gastric cancer, therapeutic strategies aimed at curbing MRPL3 activity or expression could represent a groundbreaking approach for managing this malignancy. Targeted inhibition of MRPL3 may lead to enhanced EGFR turnover, resulting in the dampening of EGFR signal transduction pathways and ultimately thwarting gastric cancer progression. The efficacy and safety of such therapeutic approaches warrant validation in clinical settings. Despite offering substantial insights into MRPL3’s involvement in gastric cancer, our study is not devoid of limitations. For example, our research predominantly utilized cell and animal models, highlighting the need for further validation of MRPL3’s prognostic relevance in a wider clinical spectrum. Moreover, the intricate molecular dynamics underpinning the MRPL3-EGFR interaction demand deeper investigation.
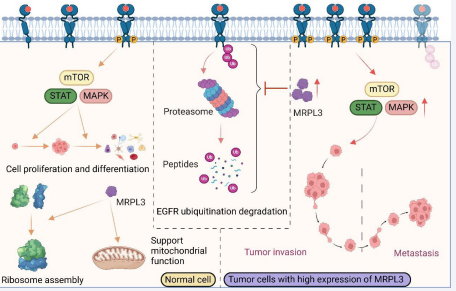
Lastly, developing small molecule inhibitors or antibody therapeutics targeting MRPL3 and assessing their clinical utility in gastric cancer treatment constitute crucial directions for future research. In summary, our investigation underscores the significant role of MRPL3 in gastric cancer progression and elucidates its function in promoting tumorigenesis through modulation of the EGFR signaling cascade [Fig 6]. These findings lay a foundational basis for the development of novel therapeutic strategies against gastric cancer.
Figure 6 Mechanism of MRPL3 in promoting gastric cancer progression. MRPL3 is a mitochondrial ribosomal protein that plays a crucial role in mitochondrial protein synthesis and energy metabolism. In the context of gastric cancer, MRPL3 is found to be significantly upregulated. This upregulation leads to the inhibition of EGFR ubiquitination, ultimately impeding the degradation process. As a result, there is an overactivation of the EGFR pathway, leading to the upregulation of downstream target genes, which in turn promotes tumor metastasis and invasion.
PATIENTS AND ETHICS
A total of 249 tissues were obtained and selected from patients with gastric cancer who underwent surgical resection at The Affiliated Hospital of Medical School, Ningbo University. Those patients were excluded from any other clinical therapies and signed informed consent forms. All studies were performed in accordance with the Declaration of Helsinki and approved by the ethics committee, [NBU-2022-054].
ACKNOWLEDGMENTS
This work was supported by the Ningbo Natural Science Foundation (No. 2022J236). We would like to express our gratitude to the Public Laboratory of Ningbo University School of Medicine for providing some of the experimental equipment.
DATA AVAILABILITY
The datasets used and/or analyzed during the current study are available from the corresponding author (Feng Gao, Gf9777@126.com) on reasonable request.
CONTRIBUTIONS
FG and CLQ conceived and designed the study. RC, SXQ, KJL, and HHS performed the cellular and molecular experiments, including assays related to MRPL3 and EGFR ubiquitination. HJ, SWQ, XHX, and TF contributed to bioinformatics analysis and statistical data interpretation. RC carried out the immunoassays related to EGFR expression. HJ participated in the design of the sequence alignment experiments. RC and HJ wrote the initial draft of the manuscript, while FG and MFZ provided critical revisions and supervised the research. All authors reviewed and approved the final version of the manuscript.
REFERENCES
- Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. GastricCancer. Lancet. 2020; 396: 635-648.
- Thrift AP, El-Serag HB. Burden of Gastric Cancer, Clin GastroenterolHepatol. 2020; 18: 534-542.
- Ajani JA, D’Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C, et al. Gastric Cancer. J Natl Compr Canc Netw. 2022; 20: 167-192.
- Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021; 71: 264-279.
- Patel TH, Cecchini M. Targeted Therapies in Advanced Gastric Cancer.Curr Treat Options Oncol. 2020; 21: 70.
- Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014; 13: 397-406.
- Cahill LS, Cameron JM, Winterburn J, Macos P, Hoggarth J, Dzamba M, et al. Structural Variant in Mitochondrial-Associated Gene (MRPL3) Induces Adult-Onset Neurodegeneration with Memory Impairment in the Mouse. J Neurosci. 2020; 40: 4576-4585.
- Guo Y, Deng X, Zhang J, Su L, Xu H, Luo Z, et al. Analysis of the MRPL3, DNAJC13 and OFCC1 variants in Chinese Han patients with TS-CTD. Neurosci Lett. 2012; 51: 18-20.
- Galmiche L, Serre V, Beinat M, Assouline Z, Lebre AS, Chretien D, et al. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum Mutat. 2011; 32: 1225-1231.
- Yin J, Lin C, Jiang M, Tang X, Xie D, Chen J, et al. CENPL, ISG20L2, LSM4, MRPL3 are four novel hub genes and may serve as diagnostic and prognostic markers in breast cancer. Sci Rep. 2021; 11: 15610.
- Gylfe AE, Katainen R, Kondelin J, Tanskanen T, Cajuso T, Hänninen U, et al. Eleven candidate susceptibility genes for common familial colorectal cancer. PLoS Genet. 2013; 9: e1003876.
- Zhang X, Dong W, Zhang J, Liu W, Yin J, Shi D, et al. A Novel Mitochondrial-Related Nuclear Gene Signature Predicts Overall Survival of Lung Adenocarcinoma Patients. Front Cell Dev Biol. 2021; 9: 740487.
- Valles I, Pajares MJ, Segura V, Guruceaga E, Gomez-Roman J, Blanco D, et al. Identification of novel deregulated RNA metabolism-related genes in non-small cell lung cancer. PLoS One. 2012; 7: e42086.
- Rashid M, Shahn SG, Verma T, Chaudhary N, Rauniyar S, Patel VB, et al. Tumor-specific overexpression of histone gene, H3C14 in gastric cancer is mediated through EGFR-FOXC1 axis. Biochim Biophys Acta Gene Regul Mech. 2021; 1864: 194703.
- 15. Zhao H, Gezi G, Tian X, Jia P, Morigen M, Fan L. Lysophosphatidic Acid-Induced EGFR Transactivation Promotes Gastric Cancer Cell DNA Replication by Stabilizing Geminin in the S Phase. Front Pharmacol. 2021; 12: 706240.
- Openshaw MR, Pinato DJ, Valeri N. Back from the Brink: EGFR Inhibition in Gastroesophageal Cancer. Clin Cancer Res. 2021; 27: 2964-2966.
- Deng J, Liang H, Zhang R, Sun D, Pan Y, Liu Y, et al. STAT3 is associated with lymph node metastasis in gastric cancer. Tumour Biol. 2013; 34: 2791-2800.
- Li SY, Hou LZ, Gao YX, Zhang NN, Fan B, Wang F. FIP-nha, a fungal immunomodulatory protein from , induces apoptosis and autophagy in human gastric cancer cells via blocking the EGFR-mediated STAT3/ Akt signaling pathway. Food Chem (Oxf). 2022; 4: 100091.
- Peng L, Jiang J, Chen HN, Zhou L, Huang Z, Qin S, et al.Redox-sensitive cyclophilin A elicits chemoresistance through realigning cellular oxidative status in colorectal cancer. Cell Rep. 2021; 37: 110069.