Phylogenetic Analysis of Rotaviruses Detected in Hospitalized Children from Karachi Pakistan Reveals only 72% Similarity with Vaccine Strains
- 1. Department of Microbiology, University of Karachi, Pakistan
- 2. Centers of Excellence in Science & Applied Technologies (CESAT), Islamabad, Pakistan
- 3. Jamil-ur-Rahman Center for Genome Research, Dr. Panjwani Center for Molecular Medicine and Drug Research, International Center for Chemical & Biological Sciences, University of Karachi 75270, Karachi, Pakistan
Abstract
Introduction: Rotavirus is a well-known universal cause of pediatric gastroenteritis. It carries the highest gastroenteritis global disease burden in pediatric age group.
Objective: Present study was conducted to examine the molecular analysis of circulating strains of rotavirus in southern Karachi and other regions of Pakistan with the global strains; and to determine their relationship with the vaccine strains to define and predict the efficacy of available vaccines.
Place and Duration of Study: Pediatrics department of PNS Shifa Hospital and Abbasi Shaheed Hospital Karachi during November 2014 to August 2016.
Study Design: It was a perspective observational study.
Methodology: Stool samples collected from hospitalized children under five years of age. Rotavirus detection was performed through ELISA. Samples were genotyped by Polymerase chain reaction. The strains of G3 and G12 were sequenced and submitted to Genbank NCBI. MK355446 and MK904566 were assigned to G12 rotavirus sequences while MK355445 and MK185023 to G3 rotavirus sequences. These sequences were aligned and matched against regional and global strains as well as with the available vaccine strains.
Results: Our G3 strain sequences were found 99-100% similar with the reference strains, and 95% similar with the G1[P8] wild type, rotarix and pentavalent reassortant Rotateq strains, while our G12 sequence was found to have 97-100% similarity with the reference sequences from the world. However, these strains are at far distance from available vaccine strains (72% similarity).
Conclusion: The G12 sequence with 72% similarity with vaccine strains raises the question for use of these vaccines in our community and becoming a cause of prolonged surveillance of this virus burden and circulating viral strain type.
Citation
Nisa TU, Ahmad A, Baber MM, Tahir A, Khan SU, et al. (2021) Phylogenetic Analysis of Rotaviruses Detected in Hospitalized Children from Karachi Pakistan Reveals only 72% Similarity with Vaccine Strains. JSM Gastroenterol Hepatol 8(1): 1103.
Keywords
• Pediatric Diarrhea
• Rotavirus genotype
• G3 rotavirus strains
• G12 rotavirus strains
INTRODUCTION
Rotavirus is the well-known cause of pediatric gastroenteritis world-wide [1]. The rotavirus, is a non-enveloped virus with 8-11 double stranded (ds) RNA segmented genome that encode 12 protein, six each structural protein (VP1-4, 6&7) and nonstructural (NSP1-6) protein. VP4 is protease sensitive defined as P type, while VP7 glycoprotein is defined as G type, both are responsible of determining serotype and genotype of the rotavirus. The most common genotype of the rotavirus around the globe are G1[P8], G2[P4], G4[P8], G9[P8] and G12[P8] & G3[P8] that are lately emerging prevalent strains [2]. Previously reported Rotavirus strain typing data of Pakistan is alarming with the % positivity (17, 29.4, 57.3, 18.9-26.3, 23.8, 34, 30.5, 63 and 23.2) respectively. The most important circulating strains are G1[P8], G2[P4],G9[P8] and G3[P8] [3-11]. Rotavirus G3 and G12 have been reported earlier from Pakistan however more data is required to get in depth knowledge for rotavirus strain diversity. G12 was first emerged in Philippine during 1987 [12]. In Pakistan, 1st G12 was reported a decade back [13].
The main objective of this study was to describe the epidemiological characteristics as well as to determine the circulating genotypes of rotaviruses in southern Karachi, Pakistan. Although the rotavirus gastroenteritis has been declared as a vaccine preventable disease and a few rotavirus vaccines are available in global market, but the disease is still prevalent. From the available vaccines, Rotarix monovalent G1P [8] and Rotateq vaccine pentavalent reassortant contains G1, G2, G3, G4, and G9, are commonly used.
The sequence of our G3 and G12 strains were aligned and matched against regional and global strains as well as with the available vaccine strains sequences. The present data envisaged to provide some relationship of the present strain diversity of Karachi region with the vaccine strains as well as other regional and global strains of Rotavirus to define and predict the efficacy of available vaccines.
MATERIALS AND METHODS
In order to gain insight into the degree of variability in the rotavirus strains circulating in Karachi Pakistan, nucleotide sequences of rotaviruses isolated from 04 representative fecal samples were compared with the reference strains. For this, the viral genome was isolated from fecal samples using the in house developed RNA/DNA extraction kit by spin column method. The quantity and quality of isolated DNA/RNA was determined using Multiskan Go spectrophotometer (Thermo Scientific, United Kingdom). Primer specific Reverse transcription PCR was performed by using primer VP7R and 9Con 1L. A semi-nested multiplex Genotyping PCR was run by using 9con1L (VP7 forward consensus primer) and a reverse primer pool containing 10 pico mole each primer 1T1, 9T1, 9T2, 9T3, 9T4, 9T9B and Mo109 for G1, G2, G3, G4, G9 and [14] G12 respectively. The detailed primer sequence and cycling condition is represented in Table 1.
PCR amplicons from successful VP7 PCR purified with GenJet PCR purification kit EU Lithuania, according to manufacturer instruction. The concentrations of amplified DNA were determined using a MultiSkan Go spectrophotometer (Thermo Scientific, United Kingdom). The purified PCR amplicons were subject to Sanger’s sequencing with Big Dye Terminator chemistry using the primer VP7R (Reverse Primer) and 9T3P and Mo109, G3 and G12 specific primer respectively. The nucleotide sequences of group A rotavirus described in the present study have been deposited in the GenBank database. For phylogenetic analysis and comparison with rotavirus sequences reported from other regions of the world, the prototype sequences of rotavirus were retrieved from the GenBank database based on the closest nucleotide sequence relatedness with the strains sequenced in the present study using BLAST search (www.ncbi.nlm.nih. gov/blast) these are indicated with their Genbank accession numbers and country names. Phylogenetic tree and analysis of VP7 nucleotide sequences of G3 and G12 genotype rotaviruses was performed by using neighbor-joining method with the 1000 replicates using the MEGA X program version 10.0.5. For G3, the optimal tree with the sum of branch length =0.25699209 while for G12, the optimal tree with the sum of branch length was 0.06883256. Percent bootstrap support is indicated by a number at each branch node. Reference strain sequences were obtained from the GenBank database and named with accession numbers and countries.
RESULTS
The nucleotide sequences of group A rotavirus described in the present study have been deposited in the GenBank database with the access numbers as follows: MK355446 and MK904566 for strain PK026 & PK0015 (G12), and MK355445 and MK185023 for strain PK004 & PK022 (G3) respectively (Table 2).
The BLAST search showed that G3 strains isolated in this study were closely related to each other as well as other previously reported Pakistani G3 MG7445621, MG744565 and MG744564 from Lahore, MG735435, MG735433 and MG735432 from Rawalpindi and MF873451 from Karachi (100% similar). Moreover, high homology (99%) was observed with another set of previously reported Pakistani strains G3 MG735439, MG735438, MG735437, MG735436, MG735434 from Rawalpindi (2015) and MG744563 and MG744566 (2016) from Lahore as well as other neighboring countries of the region like KY774435 and MF621179 from India (2016), MH171725 from Bangladesh (2016), EU708580 (2006) and EU708585 (2007) from China, MF469306, and MF469446 from USA, GU393006 (2008) from Germany and KX362760 (2013) and KX362815(2014) from UK was found. Also, our strains have shown 95% homology with the reassortant Rotateq G3 GU565079 strain sequence of pentavalent Rotateq vaccine.
Likewise, through the BLAST search of G12 strains detected in this study were found closely related (100% similar) to each other, and MF673458-9 previously reported from Karachi and MG744568 from Rawalpindi Pakistan during 2015-16. The strains KY412192 from Iran and DQ501280 from UK were 99% similar (2003), EF559260 from India 97% similar, DQ490570- 1(2003), EU839943-4 from Bangladesh (2005-6), FJ52121, KJ752009(2010) from USA, FJ747630(2008) from Germany were 97-98% similar. Our sequence was found quite distant with GU565079 strain sequence of pentavalent Rotateq vaccine, JX9436142 Rotarix vaccine strain G1P [8] and wild type G1P [8] strains (72% similar).
Table 1: The primer used for genotype with primer sequence and product size.
| PCR | Primer Name | Sequence (5' to 3') G Type | PCR Product Size (bp) |
| Primer Specific RT PCR | 9con1-L | 5’-TAG CTC CTT TTA ATG TAT GG-3’ | |
| VP7 R | 5’-GTA TAA AAT ACT TGC CAC CA-3’ | ||
| Second Round Semi-nested PCR for typing | 9T-1 (G1) | 5’-TCT TGT CAA AGC AAA TAA TG-3’ | 158 |
| 9T-2(G2) | 5’-GTT AGA AAT GAT TCT CCA CT-3’ | 244 | |
| 9T-3P(G3) | 5’-GTC CAG TTG CAG TGT AGC-3’ | 464 | |
| 9T-4(G4) | 5’-GGG TCG ATG GAA AAT TCT-3’ | 403 | |
| 9T-9B(G9) | 5’-TAT AAA GTC CAT TGC AC-3’ | 110 | |
| Mo109(G12) | 5’-TAA CGC TAA TGA ATT TTG GTA CTG-3’ | 463 |
Table 2: Strains ID with their genotype and respective gene bank accession number
| Study ID | Genotype | Genbank Accession Number Assigned |
| PK0004 | G3 | MK185023 |
| PK0022 | MK355445 | |
| PK0026 | G12 | MK904566 |
| PK0015 | MK355446 |
DISCUSSION
For determining the evolutionary footprints of viral strains under study, the phylogenetic trees were constructed from nucleotide sequences obtained from G3 MK355445 and MK185023, and G12 MK355446 and MK904566 strains. The phylogenetic trees were constructed based on the neighborjoining method with the 1000 replicates using the MEGA X program version 10.0.5 (Figure 1.
Figure 1: Phylogenetic analysis of VP7 nucleotide sequences of G3 genotype rotaviruses. G3 of our study is mentioned as ? while previously reported Pakistani G3 strains are marked as ?. The tree was constructed based on the neighbor-joining method with the 1000 replicates using the MEGA X program version 10.0.5. The optimal tree with the sum of branch length =0.25699209 is shown for G3 and. Percent bootstrap support is indicated by a number at each branch node. Reference strain sequences were obtained from the GenBank database and named with accession numbers and countries. The variation scale for G3 is indicated as 0.02. G3 strains Genbank accession numbers are (MK185023 and MK355445) for strain PK0004 & PK0022 respectively.
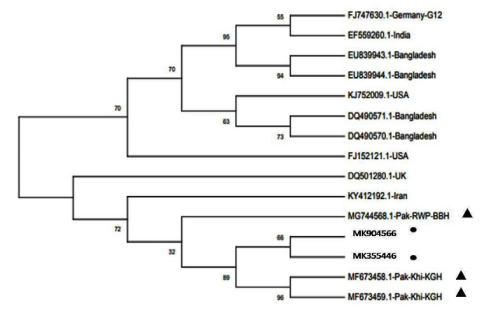
Figure 2.
Figure 2: Phylogenetic analysis of VP7 nucleotide sequences of G12 genotype rotaviruses. G12 of our study mentioned as ? while previously reported Pakistani strains are marked as ?. The tree was constructed based on the neighbor-joining method with the 1000 replicates using the MEGA X program version 10.0.5. The optimal tree with the sum of branch length =0.06883256 is shown for G12. Percent bootstrap support is indicated by a number at each branch node. Reference strain sequences were obtained from the GenBank database and named with accession numbers and countries. The variation scale for G3 is indicated as 0.005. G12 strains Genbank accession numbers are (MK355446 and MK904566) for strain. PK0015 &PK0026 respectively.
Phylogenetic study of our reported nucleotide sequence of G3 strains with the other local, regional and global strains is indicative of the similar lineage of the rotavirus G3. The highest homology (99-100%) of our reported sequence of G3 strains with other early reported sequences from Pakistan, South Asia, other countries of the world as well as with the vaccine strains could be due to two reasons: One that our reported sequence is smaller in size; secondly there may be some prominent differences if we had a bigger data set or the larger nucleotide sequence from the same gene used for analysis. Despite of the above mentioned fact, the similarity of our reported sequences of G3 rotavirus with the earlier reported Pakistani strains and vaccine strains (95% similarity) and presence of G3 rotavirus with high prevalence number (22.9%) in another data set from north of Pakistan during similar period of study [11], strongly suggests the requirement of implication of rotavirus vaccine in our region,.
The similarity of G12 strains in both studies shows that these are from the same lineage like with other previously reported Pakistani strains from south as well as from north during same era as of our study. These are also presented as sub lineage of Germany, USA, Bangladesh and is the first Indian reported G12 strains with 97% similarity. In Bangladesh during 2012-2015, a hospital based surveillance revealed the 29% prevalence of G12 [P8] [15], two reports from Indian cities also point towards the presence of G12 as 14.8% & 15.6% during 2007-2012 [16] and [17], even from Pakistan the previous report indicated the 19.8% prevalence of G12 strains [18,19] advocates the presence of G12 in high number in south Asia region. Secondly in our data set, the distance of almost all G12 strains with the wild type G1[P8] rotavirus, GU565079 strain sequence of pentavalent Rotateq vaccine and JX9436142 Rotarix vaccine strain G1P [8] leaves a question about the use of these available vaccines and the cross protection against rotavirus G12 in children of this region.
CONCLUSION
This study demonstrates the presence of globally common G3 and G12 strains of rotavirus in southern Karachi, Pakistan It requires constant surveillance at national level to determine disease burden and genotype data. Furthermore, efficacy of available vaccines may be determined to recommend using in Pakistani community. Emergence of any new rotavirus strain type must also be monitored which may facilitate the relevance of existing vaccines and consideration of new vaccine candidates.
ETHICAL APPROVAL
Study was approved from the Institutional Ethical Review Committee.
DISCLAIMER
The paper is the part of PhD thesis of author Tayyab Un Nisa
ACKNOWLEDGEMENT
We are thankful to Dr. Imran Ahmad, Dr. Rida, Dr. Shamsa Aqeel and Dr. Sunbul Waheed for the provision of stool samples. We thank Dr. Rakhshanda Bilal for her guidance related to research work and effort of liaison for sample collection. Mr. Hamid Ahmed for providing assistance during lab experiments.
SOURCE OF FUNDING
Institutional indigenous scholarship for Post Graduate studies