Selection of Leucine as a Potential Antagonist from In Silico Analysis of -Opioid Receptor in the Treatment of Subjects with Heroin and Opiate Addiction
- 1. Department of Human Anatomy, Ahmadu Bello University, Nigeria
- 2. Department of Biochemistry, University of Ilorin, Nigeria
- 3. Department of Human Anatomy, University of Ilorin, Nigeria
- 4. Department of Psychology, Concordia University, Canada
ABSTRACT
The aim of this study was to analyze the antagonistic potential of leucine on μ-opioid receptor by computational studies. Studies has shown that drug addiction has reached epidemic levels across the globe with approximately 247 million drug users worldwide. Heroin binds to and stimulate the μ-opioid receptor thereby inducing the release of neurotransmitter dopamine, triggering excessive drug taking behavior. The life-threatening side effects of the current μ-opioid receptor drugs (Suboxone and Naloxone) such as Asthenia, Insomnia, Rhinitis, Infections, Pain, Headache e.t.c necessitate the discovery of novel potent and safe compounds as a therapeutic approach in the treatment of drug addiction. In view of this, computational tools were adopted to out-source better antagonist for μ-opioid receptor as a drug-gable target. The Leucine chemical compound was retrieved from PubChem database and was screened for its inhibitory potential on μ-opioid receptor, which was retrieved from protein data bank repository. PyRx AutoDock Vina option based on scoring functions was used to perform Computational docking analysis and the target was validated in order to ensure that the right target and appropriate docking protocol was used for this study. Leucine was found to have a better binding affinity with the target (-4.7kcal/mol) when compared with the co-crystallized molecule (-2.5kcal/mol). Leucine has a molecular weight (MW) of 131.174 g/mol, number of hydrogen bond donor is 2, number of hydrogen bond acceptor is 3, LogP is -1.864 and number of rotatable bond is 3. Docking studies and ADME/T (Absorption, Distribution, Metabolism, Excretion and Toxicity) properties of leucine on μ-opioid showed that this ligand is a druggable molecule when docked well with μ-opioid. Therefore, Leucine plays an inhibitory role on μ-opioid receptor and thus should be implicated as a potential agent in the treatment of drug addiction.
KEYWORDS
- Dopamine
- Naloxone
- PyRx AutoDock Vina
- Suboxone and Toxicity
CITATION
Oladimeji O, Ambrose GO, Umana U, Olayemi O, Abel A, et al. (2024) Selection of Leucine as a Potential Antagonist from In Silico Analysis of µ-Opioid Receptor in the Treatment of Subjects with Heroin and Opiate Addiction. J Addict Med Ther 11(1): 1048.
INTRODUCTION
Opiate drugs are widely used as analgesic to induce antinociception and to treat pain disorders [1-3]. However, the absued use of opioids results in a conceptualized three- stage circle of binge/intoxication, withdrawal/negative affect, and preoccupation/anticipation that reflects dysregulation in three functional domains (incentive salience/habits, negative emotional states, and executive function, respectively) of the brain [4]. This effect worsens over time and involves neuroplastic changes in the brain regions involved in reward, stress, and executive functions [5]. The growing availability of low-cost synthetic opioids, such as non-pharmaceutical fentanyl’s has played a profound role in fostering this endemic crisis among drug users [6].
The early 1970s recorded a notable discovery that opiate drugs bind to receptors in the brain and employs a complex endogenous neuromodulatory system to exercise their pharmacological effects [7]. The opioid system comprises three homologous G protein- coupled receptors (GPCRs) known as mu-, kappa- and delta opioid receptors (MORs, KORs and DORs respectively [8]. These subtypes share a common analgesic effect in brain, and each has their unique effects such as MOR for euphoria and respiratory depression, KOR for dysphoria, and DOR opioid receptor for anxiolysis [3]. Opioid receptors are activated by endogenous opioid peptides, forming a peptide family that includes β- endorphin, enkephalins and dynorphins [9].
MOR was the first opioid receptor discovered for its antagonistic activity that could theoretically cause euphoria, making it extremely crucial for highly complex brain reward circuits [10]. MOR are highly concentrated in brain regions that are part of the pain and reward networks (ventral tegmental area, nucleus accumbens, and cortex) which accounts for its strong reinforcing effects, euphoria and the inducing properties of rewarding stimuli, playing an important role in goal-directed behavior such as drug-seeking behavior for pleasure [11]. In addition, MORs are expressed in brainstem regions that regulate breathing which accounts for its agonistic action in respiratory depression, which is the primary cause of death [12].
Opioids binds to the MOR, to send signals to the dopamine terminal, resulting in a large increase in the release of dopamine by dopaminergic neurons in the tergmental area (by inhibiting γ-aminobutyric acid (GABA)). Dopamine increase in this circuit reinforces drug-seeking behaviour, which essentially teaches the brain to repeat the action in opioid addiction [13]. Thus, this demonstrate MOR has a the key responsible receptor for the adverse actions of opioid agonist.
Branched Chain Amino Acids (BCAAs) plays an important role in brain function by regulating brain protein synthesis and production of energy. BCAAs influence synthesis of different neurotransmitters, which includes; dopamine, serotonin and norepinephrine directly or indirectly [14]. Leucine belongs to this group of BCAAs, (3 isoleucine (ILE), and valine (VAL)) which participate in a variety of important biochemical functions in the brain and has been examined as a potential supplement in the treatment for several neurological diseases [15,16]. Leucine can be predominantly found in Animal foods: Eggs, Dairy, Meat (Chicken and Fish) and plant foods [17].
Administration of leucine as a competing neutral amino acid, increases BCAAs plasma concentration and brain absorption of BCAAs which ultimately, leads to a decrease in the rate of conversion to dihydroxyphenylalanine (DOPA) to Dopamine and hence, regulates the synthesis of this neurotransmitter [18]. It is expected that targeting the reduction in the release of dopamine through MOR inhibition could be highly beneficial in the treatment of opioid drug reinforcement. The purpose of this study was to ultilize computational approach to investigate the interaction between dietary leucine supplementation and MOR as a novel approach towards preventing drug seeking behaviour and reinforcement in opioid addiction.
Lipinski rule of five on ADMET (Adsorption, Distribution, Metabolism, Excretion and Toxicity) properties was used to evaluate the Leucine ligand an d it was found to fulfill the rule of five on ADMET properties.
METHODOLOGY
Ligand selection and Preparation
The chemical structures of Leucine (PubChem ID: 6106) was obtained from PubChem compound database (https://pubchem. ncbi.nlm.nih.gov) [19]. The MOL SDF format of this ligands was converted to PDBQT file using PyRx tool to generate atomic coordinates and energy was minimized by optimization using the optimization algorithm at force field set at uff (required) on PyRx.
Accession and Preparation of the Target Protein
The protein Mu-receptor was prepared by retrieving the three dimension crystal structure of mu-receptor in complex with a co-crystallized ligand (PDB: 6DDE) from RCSB PDB (http://www.rcsb.org/pdb/home/home.do) [20]. The protein was subsequently cleaned by using Pymol tool to remove the bound complex molecule, non-essential water molecules and all the heteroatoms. The co-crystallized ligand (PRD_002308) was extracted (not removed) from the active site so as to reveal the grid coordinate around the binding pocket when viewed on Pymol.
Molecular docking using PyRx
After the receptor and ligands were prepared, PyRx, AutoDockVina option based on scoring functions was used to perform molecular docking analysis. For our analysis we used the PyRx, Auto Dock Vina exhaustive search docking function. After the minimization process, the grid box resolution was centered at 1.878, 15.7749, -48.3753 along the x, y and z axes respectively at grid dimension of 25x 25 x 25 Å to define the binding site. The co- crystallized ligand which serves as the standard was first docked within the binding site of MOR and the resulting interaction was compared with that of lecuine into the related active sites with the same grid box dimension.
RESULT AND DISCUSSION
In the present study, Leucine was docked into the binding pocket of MOR for its (antagonistic) properties. Leucine was discovered as the lead compound with the energy of -4.7 kcal/ mol when compared with the co-crystalized ligand [Table 1].
Table 1: Docking scores and RMSD values of Ligands.
|
Ligand |
Binding Affinity (kcal/mol) |
RMSD/UD |
RMSD/LD |
|
Buprenorphine |
-4.9 |
0 |
0 |
|
Leucine |
-4.7 |
0 |
0 |
|
Co-crystalized Ligand |
-2.5 |
0 |
0 |
*RMSD/UB: Root mean square deviation/upper bond; RMSD/LB: Root mean square deviation/lower bond.
Table 2: Lipinski's drug-like properties of Leucine: The rule describes drug candidate’s pharmacokinetics in the human body which also including their absorption, distribution, metabolism, and excretion
|
Molecular Properties |
Lipinski’s rule of Five |
Leucine’s drug-like properties |
|
Molecular Mass g/Mol |
<500 |
131.174 |
|
Hydrogen bond Acceptor |
<10 |
3 |
|
Hydrogen bond donor |
<5 |
2 |
|
LogP |
<5 |
1.864 |
|
No of rotatable bond |
<5 |
3 |
(“ADME/T’) using an online server. (http://www.scfbio-iitd.res.in/).
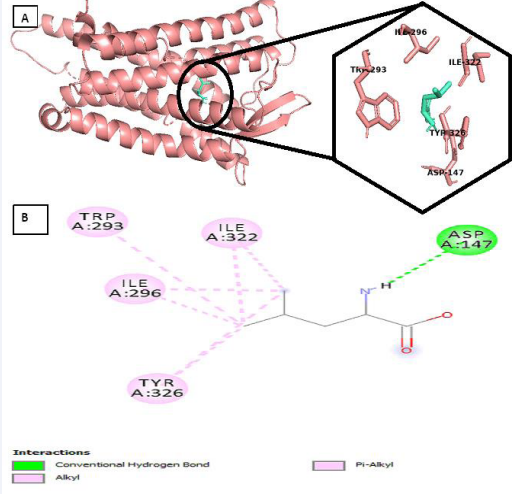
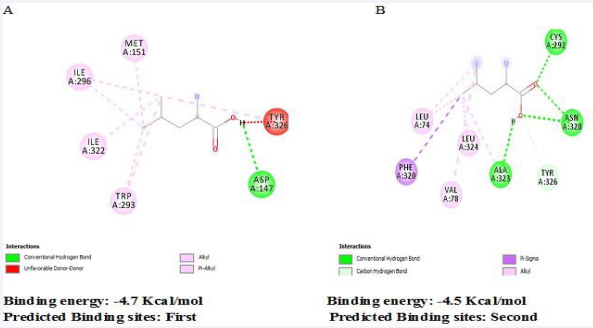
The drug-likeness of Leucine was assessed by subjecting it to the Lipinski’s rule of five, afterwards the lead compound, Leucine violated none of the rules, and this describes its bioavailability and binding potential [Table 2]. The high binding energy (-4.7 Kcal/ Mol) attributed to Leucine when compared to the co crystalized ligand (-2.5 Kcal/Mol) in this regard is believed to be as a result of its chemical interaction at the receptor active site [Table 3; Figure 2], which includes one (1) Conventional hydrogen bond involving A-147 residues; six (6) Hydrophobic interaction involving A-326, A-293, A-296 and A-322 residues.
Figure 2 (a) Pose view of leucine at optimum binding (b) 2D interactions of the leucine within the binding pocket.
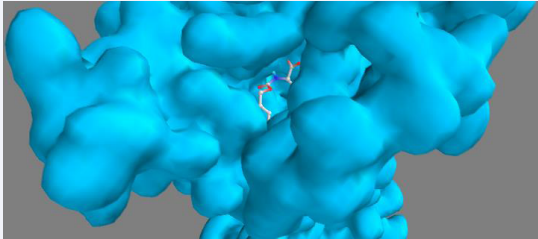
Figure 3 Leucine within the binding pocket.
Table 3: Interaction table showing the various chemical interactions of Leucine within the binding pocket (Viewed on Discovery studio Visualizer)
|
S/N |
Name |
Category |
Type |
|
1 |
N:UNK1:H - A:ASP147:OD2 |
Hydrogen Bond |
Conventional Hydrogen Bond |
|
2 |
N:UNK1:C - A:ILE296 |
Hydrophobic |
Alkyl |
|
3 |
N:UNK1:C - A:ILE322 |
Hydrophobic |
Alkyl |
|
4 |
N:UNK1:C - A:ILE296 |
Hydrophobic |
Alkyl |
|
5 |
N:UNK1:C - A:ILE322 |
Hydrophobic |
Alkyl |
|
6 |
A:TRP293 - N:UNK1:C |
Hydrophobic |
Pi-Alkyl |
|
7 |
A:TYR326 - N:UNK1:C |
Hydrophobic |
Pi-Alkyl |
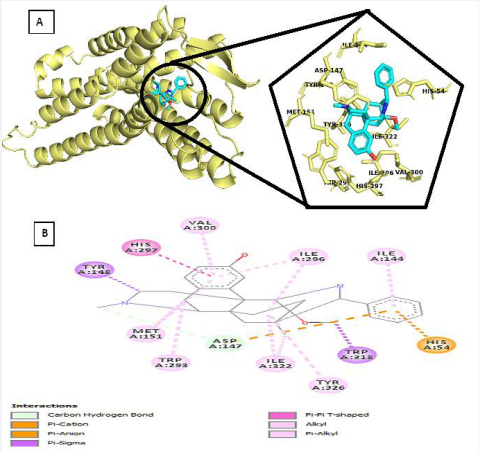
While that of the co-crystalized ligand which serves as standard presents with the following chemical interaction at the binding pocket [Table 4; Figure 1]. One (1) Carbon Hydrogen bond involving A-147; Two (2) Electrostatic interaction involving A-54 and A-147 residue; and fifteen (15) hydrophobic interactions.
Figure 1 (a) Pose view of Co-crystakized ligand at optimum binding (b) 2D interactions of the co-crystalized ligand within the binding pocket.
Table 4: Interaction table showing the chemical interactions of the co-crystalized ligand within the binding pocket (Viewed on Discovery studio Visualizer)
|
S/N |
Name |
Category |
Type |
|
1 |
N:UNK1:C - A:ASP147:O |
Hydrogen Bond |
Carbon Hydrogen Bond |
|
2 |
A:HIS54:NE2 - N:UNK1 |
Electrostatic |
Pi-Cation |
|
3 |
A:ASP147:OD2 - N:UNK1 |
Electrostatic |
Pi-Anion |
|
4 |
N:UNK1:C - A:TYR148 |
Hydrophobic |
Pi-Sigma |
|
5 |
N:UNK1:C - A:TRP318 |
Hydrophobic |
Pi-Sigma |
|
6 |
A:HIS54 - N:UNK1 |
Hydrophobic |
Pi-Pi-T-shaped |
|
7 |
A:HIS297 - N:UNK1 |
Hydrophobic |
Pi-Pi-T-shaped |
|
8 |
A:MET151 - N:UNK1 |
Hydrophobic |
Alkyl |
|
9 |
N:UNK1:C - A:ILE322 |
Hydrophobic |
Alkyl |
|
10 |
N:UNK1 - A:ILE296 |
Hydrophobic |
Alkyl |
|
11 |
N:UNK1 - A:ILE322 |
Hydrophobic |
Alkyl |
|
12 |
N:UNK1:C - A:MET151 |
Hydrophobic |
Alkyl |
|
13 |
A:TRP293 - N:UNK1:C |
Hydrophobic |
Pi-Alkyl |
|
14 |
A:TYR326 - N:UNK1 |
Hydrophobic |
Pi-Alkyl |
|
15 |
N:UNK1 - A:MET151 |
Hydrophobic |
Pi-Alkyl |
|
16 |
N:UNK1 - A:ILE296 |
Hydrophobic |
Pi-Alkyl |
|
17 |
N:UNK1 - A:VAL300 |
Hydrophobic |
Pi-Alkyl |
|
18 |
N:UNK1 - A:ILE144 |
Hydrophobic |
Pi-Alkyl |
Hydrogen (H)-bonds potentiates diverse cellular cellular functions by facilitating molecular interactions. In order words, hydrogen bonds are considered to be facilitators of protein-ligand binding [21]. Previous studies have shown that synergistics receptor-ligand H-bond pairing potentiate high-affinity binding which corresponds to an increase in binding affinity [22]. It is obvious that the higher binding affinity of Leucine to the binding pocket of the MOR when compared to that of the co-crystalized ligand is attributed to the presence of “strong” conventional hydrogen bond present in Leucine when compared to the standard which has the “weak” carbon hydrogen bond [23].
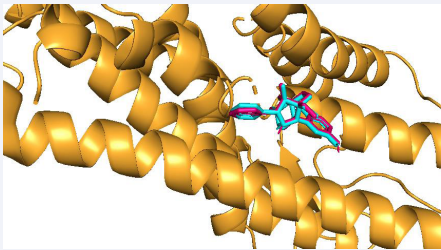
We validated the accuracy of our docking protocol by redocking the co-crystalized ligand back into the binding pocket of the MOR. As stated, the redocked pose overlapped almost totally with the experimental orientation, indicating that Auto dock vina on PyRx re-docked the co-crystalized ligand, with a very high accuracy, back into the binding pocket of the MOR. This shows that our method of docking was reliable and the resulting docking scores are correct [Figure 4].
Figure 4 Validation of docking: Comparability of the re-docked binding mode of co-crystalized ligand within MOR binding pocket. A snapshot from PyRx.
BINDING SITE PREDICTION AND BINDING MODE ANALYSIS
Based on the Meta Pocket 2.0 server, we were able to identify three potential binding sites capable of accommodating the ligands with varying binding affinities [Figures 5].
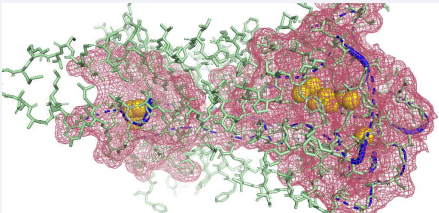
Figure 5 The result of residue mapping of mu-receptor. Image generated from PyMOL 1.2. PDBID: 6DDE. Ligand binding sites are illustracted in yellow ball, potential binding atoms are in blue cartoon, functional residues are in red mesh, other parts of the protein are in green sticks.
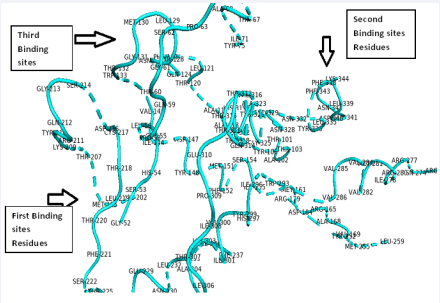
This suggests that leucine binds to mu-receptor with varied binding affinities based on its binding modes or orientation of binding with respect to the respective binding sites. The three sites with their respective amino acids residues are located in the monomer [Figure 6].
Figure 6 The functional amino acids residues for the three predicted binding sites.
As can be observed in [Figure 7], Leucine associate with the highest binding affinity with amino acids predicted in binding pocket 1 such as MET151, ILE296, ILE322, TRP293, ASP147 and TRP326 a lower binding affinity with binding pocket 2. This reveals that the activity of mu-receptor is better regulated at the predicted binding pocket 1 and not 2.
Figure 7 Predicted binding sites by MetaPocket 2.0 sever (a) Pocket 1 (b) Pocket 2.
CONCLUSION
Docking studies and ADMET evaluation of Leucine with MOR showed that this ligand is a drug-gable molecule, which docks well with MOR. Therefore, Leucine molecule plays an important role in inhibiting MOR and thus should be implicated as a potential agent in substance abuse disorder.
ACKNOWLEDGMENT
The Authors highly acknowledge the support of all staff and students of the department of Anatomy, Ahmadu Bello University, Zaria, Nigeria.
REFERENCES
- Colvin LA, Bull F, Hales TG. Perioperative opioid analgesia-When is enough too much? A review of opioid-induced tolerance and hyperalgesia. Lancet. 2019; 393: 1558-1568.
- Wang SC, Chen YC, Lee CH, Cheng CM. Opioid Addiction, Genetic Susceptibility, and Medical Treatments: A Review. Int J Mol Sci. 2019; 20: 4294.
- Seth P, Scholl L, Rudd RA, Bacon S. Overdose deaths involving opioids, cocaine, and psychostimulants-United States, 2015-2016. MMWR Morb Mortal Wkly Rep. 2018; 67: 349-358.
- Koob GF, Volkow ND. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 2018; 3: 760-773.
- Goldstein RZ, Volkow ND. Drug addiction and its underlying neurobiological basis: neuroimaging evidence for the involvement of the frontal cortex. Am J Psychiatry. 2002; 159: 1642-1652.
- Le Merrer J, Becker JA, Befort K, Kieffer BL. Reward processing by the opioid system in the brain. Physiol Rev. 2009; 89: 1379-1412.
- Brownstein MJ. A brief history of opiates, opioid peptides, and opioid receptors. Proc Natl Acad Sci. 1993; 90: 5391-5393.
- Volkow D, Jones B, Einstein B, Wargo M. Prevention and Treatment of Opioid Misuse and Addiction. JAMA Psychiatry. 2019; 76: 208-216.
- Le Merrer J, Becker JA, Befort K, Kieffer BL. Reward Processing by the Opioid System in the Brain. Physiol Review. 2009; 89: 1379-1412.
- Contet C, Kieffer BL, Befort K. Mu opioid receptor: A gateway to drug addiction. Curr Opin Neurobiol. 2004; 14: 370-378.
- Ugur M, Derouiche L, Massotte D. Heteromerization Modulates mu Opioid Receptor Functional Properties in vivo. Front Pharmacol. 2018; 9: 1240.
- Imam MZ, Kuo A, Ghassabian S, Smith MT. Progress in understanding mechanisms of opioid-induced gastrointestinal adverse effects and respiratory depression. Neuropharmacol. 2018; 131: 238-255.
- Leventelis C, Goutzourelas N, Kortsinidou A, Spanidis Y, Toulia G, Kampitsi A, et al. Buprenorphine and Methadone as Opioid Maintenance Treatments for Heroin-Addicted Patients Induce Oxidative Stress in Blood. Oxid Med Cell Longev. 2019; 2019: 6417048.
- Fernstrom JD, Fernstrom MH. Tyrosine, Phenylalanine, and Catecholamine Synthesis and Function in the brain. J Nutr. 2007; 137: 1539S-1547S.
- Fernstrom JD. Branched-chain amino acids and brain function. J Nutr. 2005; 135: 1539S-1546S.
- Blomstrand E. A role for branched-chain amino acids in reducing central fatigue. J Nutr. 2006; 136: 544S-547S.
- Górska-Warsewicz H, Laskowski W, Kulykovets O, Kudli?ska-ChylakA, Czeczotko M, Rejman K. Food Products as Sources of Protein and Amino Acids-The Case of Poland. Nutrients. 2018; 10: 1977.
- Monirujjaman Md, Ferdouse A. Metabolic and Physiological Roles of Branched-Chain Amino Acids. Advan Mol Biol. 2014.
- https://pubchem.ncbi.nlm.nih.gov/
- https://www.rcsb.org/
- Salentin S, Schreiber S, Haupt J, Adasme F, Schroeder M. Molecular Biology. 2014; 116: 174.
- Sawada et al. J Am Chem Soc. 2010; 132: 16862.
- Desiraju G, Steiner T. The weak Hydrogen Bond: In structural Chemistry and Biology. Int Union Crsytallography. 2001.