Development of an Automated Diagnostic System Using Genetic Algorithms in Electroneurodiagnostic Data
- 1. National Technological Institute of Mexico, Mexico
Abstract
This project aims to develop an automated diagnostic system that leverages genetic algorithms (GAs) for analyzing electroneurodiagnostic (END) data, including electroencephalograms (EEG) and electromyograms (EMG). The growing complexity of END data poses significant challenges for accurate diagnosis and timely intervention in neurological disorders. By utilizing genetic algorithms, we aim to enhance the feature selection process, optimizing the identification of relevant patterns associated with various neurological conditions. The system will undergo rigorous training using a diverse dataset, allowing it to recognize and classify abnormalities effectively. Initial results indicate that GAs can significantly improve diagnostic accuracy compared to traditional methods, reducing the likelihood of misdiagnosis and facilitating early intervention. The project also aims to establish a user-friendly interface for clinicians, enabling them to interpret results intuitively. This innovative approach enhances diagnostic capabilities and contributes to neuroinformatics, promoting the integration of artificial intelligence in clinical practice.
Keywords
• Genetic Algorithm; Electroneurodiagnostic; Classification; Feature Reduction; Automated Diagnosis
Citation
Marquez BY, Quezada A, Alanis A, Magdaleno-Palencia JS (2025) Development of an Automated Diagnostic System Using Genetic Algorithms in Electroneurodiagnostic Data. J Neurol Disord Stroke 12(2): 1236.
INTRODUCTION
Neurological disorders are among the leading causes of disability worldwide, with conditions such as epilepsy [1]. Accurate diagnosis and timely intervention are critical to improving patient outcomes, yet these remain significant challenges in clinical practice [2,3]. The complexity and volume of electroneurodiagnostic (END) data, such as electroencephalograms (EEG) and electromyograms (EMG), add layers of difficulty for healthcare professionals, requiring both specialized expertise and advanced computational tools [4].
Traditional diagnostic methods often rely on manual analysis, which is time- consuming and prone to human error. For instance, an experienced neurologist may need hours to interpret subtle patterns in EEG data to confirm a diagnosis of epilepsy. Similarly, analyzing EMG data for neuromuscular disorders involves detecting faint anomalies that may not be easily distinguishable. As a result, an urgent need is for automated systems capable of efficiently processing and interpreting END data with high accuracy [4,5]. In recent years, artificial intelligence (AI) and machine learning (ML) have emerged as transformative tools in medical diagnostics [6]. These technologies can analyze vast amounts of data, uncovering patterns and correlations often invisible to human observers. Among AI methodologies, genetic algorithms (GAs) offer a particularly compelling approach for optimizing the diagnostic process. Inspired by the principles of natural selection, GAs is highly effective in identifying the most relevant features within complex datasets, making them an ideal choice for END data analysis [7].
The application of GAs to END data presents numerous opportunities. First, GAs can address the issue of high dimensionality by selecting only the most pertinent features for diagnosis. This not only reduces computational complexity but also enhances the interpretability of results. Second, GAs is inherently flexible, allowing them to adapt to diverse types of neurological data and conditions [8]. By integrating GAs into an automated diagnostic system, clinicians can benefit from a tool that provides real-time insights, reducing the burden of manual analysis and improving diagnostic accuracy [9]. Despite these advantages, implementing GAs in clinical settings takes time and effort. For instance, the performance of GAs heavily depends on the quality and diversity of the training dataset. More accurate data must be needed to avoid suboptimal results, underscoring the importance of rigorous dataset curation. Additionally, integrating such systems into clinical workflows requires careful consideration of usability and interpretability, as the goal is to assist clinicians rather than replace them [10].
This chapter explores the development of an automated diagnostic system that utilizes genetic algorithms to analyze END data. The proposed system aims to enhance the diagnostic process by addressing key challenges, including feature selection, noise reduction, and classification accuracy. By leveraging a robust dataset and state-of-the-art computational techniques, the system is designed to classify neurological abnormalities effectively and present results in an accessible manner for clinicians. The following sections provide an overview of the theoretical foundations of genetic algorithms and their relevance to END data analysis. The chapter also details the methodology employed in developing the diagnostic system, discusses the results of initial experiments, and highlights the implications for clinical practice and future research. Through this work, we aim to demonstrate the potential of GAs to revolutionize the field of neurodiagnostics, paving the way for more accurate and efficient medical interventions.
BACKGROUND
The background for this project integrates foundational knowledge about electroneurodiagnostic data, the specific challenges involved in its analysis, and the potential of genetic algorithms (GAs) to overcome these challenges. This section provides context for understanding the relevance of GAs in developing an automated diagnostic system and establishes the groundwork for the methodologies discussed in subsequent sections.
Electroneurodiagnostic Data
Electroneurodiagnostic (END) tests are crucial in diagnosing and managing neurological disorders. Two of the most used modalities in END are electroencephalograms (EEG) and electromyograms (EMG) [11].
EEG: This test monitors electrical activity in the brain by placing electrodes on the scalp. It is primarily used to detect abnormalities associated with epilepsy, sleep disorders, brain injuries, and other conditions. EEG data is characterized by its dynamic and complex waveforms, including alpha, beta, delta, and theta waves, which are analyzed to detect irregularities [12].
EMG: This diagnostic tool measures the electrical activity of muscles in response to nerve stimulation. EMGs are essential for diagnosing neuromuscular disorders such as amyotrophic lateral sclerosis (ALS), myasthenia gravis, and muscular dystrophy. The data typically includes bursts of electrical signals indicating muscle response under voluntary and involuntary conditions [13]. END data is inherently high-dimensional and noisy, requiring careful preprocessing to extract meaningful insights. While traditionally analyzed manually by neurologists, the increasing availability of large datasets calls for automated solutions to streamline the diagnostic process [14].
Challenges in END Analysis
Analyzing END data involves overcoming several critical challenges, which this project seeks to address through the implementation of GAs:
High Dimensionality: END data comprises numerous features, including frequency bands, amplitudes, and temporal dynamics. For example, an EEG recording can contain hundreds of data points per second across multiple channels. This sheer volume of information makes manual analysis impractical and error prone. Selecting the most relevant features from such data is a nontrivial task [15]. Neurophysiological diagnostic signals (END) are often contaminated by noise from various sources, including electrical interference, muscle movements, and environmental factors. For instance, an electroencephalogram (EEG) signal can contain artifacts from blinking or head movements, requiring effective preprocessing to filter out this noise without losing critical diagnostic information. Additionally, many neurological disorders manifest as subtle changes in END signals, such as epileptic seizures, which may be indicated by specific waveform spikes occurring sporadically. Detecting such patterns demands sophisticated analytical techniques to distinguish pathological signals from normal variations. Another significant challenge is inter-patient variability, as differences in anatomy, physiology, and the manifestation of neurological conditions can lead to substantial variations in END data across patients. Therefore, a diagnostic system must be robust enough to generalize across diverse populations while maintaining sensitivity to individualspecific anomalies [16,17].
Genetic Algorithms
Genetic algorithms (GAs) are a subset of evolutionary algorithms inspired by the principles of natural selection. They are particularly effective for optimization problems and are widely used in fields ranging from engineering to bioinformatics.
Core Principles of GAs: In genetic algorithms (GAs), the process begins with selecting candidate solutions, which are represented as chromosomes, and these solutions are evaluated based on a fitness function to determine their performance. The best-performing solutions are then chosen for reproduction, ensuring that the most promising candidates are passed on to the next generation. During crossover, pairs of selected solutions combine to produce offspring, exchanging genetic information to explore new regions of the solution space, which allows the algorithm to search for better solutions. Additionally, mutation introduces small random changes to offspring solutions to maintain diversity within the population and prevent premature convergence. This ensures the search space remains wide, and the algorithm is not stuck in suboptimal solutions.
This iterative process of selection, crossover, and mutation is particularly relevant to feature selection in END (electro-neurodiagnostic) analysis, where the goal is to reduce the dimensionality of the data while improving classification accuracy [18]. Genetic algorithms are particularly well-suited for this task, as they iteratively refine a population of feature subsets, converging toward an optimal solution that strikes the right balance between relevant and redundant features, ultimately enhancing diagnostic systems’ efficiency and accuracy [19]. Genetic algorithms (GAs) offer several advantages when applied to END (electro- neuro diagnostic) data analysis, making them highly suitable for tackling the complexities inherent in such data. One of their key strengths is adaptability, as GAs are capable of handling nonlinear relationships and intricate feature interactions, which are commonly found in END data. This adaptability enables GAs to capture complex patterns that traditional methods may miss [20]. Furthermore, GAs are scalable, meaning they can efficiently process large datasets without being hindered by the computational limitations that typically affect conventional approaches.
This scalability makes them ideal for real-world applications where the volume of data can be vast and diverse. Additionally, GAs are robust and resilient to noise and missing data, which are frequent challenges in clinical environments where data quality can vary, making them particularly effective for practical use in clinical diagnostics. Although GAs have been successfully applied in various biomedical fields, such as gene expression analysis, image segmentation, and disease risk prediction, their application to END data analysis still needs to be explored. This opens an exciting opportunity for innovation, as integrating GAs into END analysis could lead to more accurate and efficient diagnostic systems, paving the way for advancements in personalized medicine and neurological disorder detection [21].
METHODOLOGY
The methodology for developing the automated diagnostic system involves a structured approach combining data preprocessing, feature selection, classification, and user interface design. This section provides a detailed description of the system’s architecture, the role of genetic algorithms (GAs) in feature selection, the design of the classification model, and the training and validation process.
System Architecture
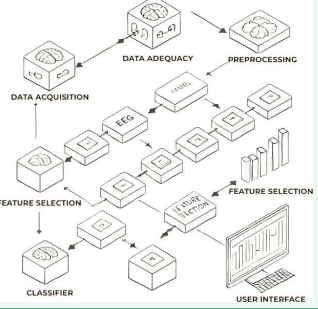
The proposed system is designed as a multi-stage pipeline, integrating several components to analyze electroneurodiagnostic (END) data effectively: The system for analyzing END (electro-neuro diagnostic) data starts with data acquisition from various sources, including publicly available datasets, clinical studies, and hospital archives, which provide EEG and EMG recordings collected under controlled conditions. However, raw END data is often contaminated by noise and artifacts, so advanced signal processing techniques, such as bandpass filtering, independent component analysis (ICA), and wavelet transformation, are applied to filter out unwanted noise while preserving diagnostically relevant features. After noise filtering, the data is segmented into more minor, manageable epochs labeled according to clinical diagnoses to create a robust training dataset. Feature extraction follows, where relevant characteristics of the underlying signals are identified; for EEG data, these include power spectral density (PSD), coherence, and frequency band powers like alpha, beta, delta, and theta waves. In contrast, features like root mean square (RMS), mean frequency, and signal entropy are computed for EMG data. A genetic algorithm (GA) is used to optimize feature selection, which iteratively refines the population of feature subsets to identify the most informative features, reducing dimensionality and improving classification accuracy [22]. A machine learning classifier, such as support vector machines (SVM), convolutional neural networks (CNNs), or ensemble methods like random forests, is then trained using the selected features to recognize patterns indicative of specific neurological conditions [23,24]. Finally, a clinician-friendly user interface is developed to present the diagnostic results, which includes visualizations like spectrograms, annotated waveforms, and classification outputs, allowing clinicians to interpret the results and make informed decisions intuitively (Figure 1).
Figure 1 System Architecture
Feature Selection Using Genetic Algorithms
Feature selection plays a pivotal role in enhancing the performance of a system, as it directly influences both computational complexity and classification accuracy, particularly in high-dimensional and noisy data. Genetic algorithms (GAs) are well- suited for this task because of their ability to efficiently search through vast feature spaces and handle complex, noisy data. The process begins with initialization, generating a population of chromosomes, each representing a subset of features. These chromosomes are encoded as binary strings, with each bit indicating the inclusion (1) or exclusion (0) of a particular feature. Next, the fitness of each chromosome is evaluated using a predefined fitness function that considers classification accuracy, computational efficiency, and feature redundancy, ensuring that the selected features contribute to a model that is both accurate and efficient. The top-performing chromosomes, those that achieve the highest fitness scores, are then chosen through roulette wheel selection or tournament selection, which also helps maintain diversity in the population. To explore new regions of the solution space and encourage genetic diversity, selected chromosomes undergo crossover, exchanging genetic material to produce offspring. At the same time, mutation introduces random changes to the offspring chromosomes. This process helps prevent premature convergence and enhances the algorithm’s ability to explore various combinations of features. Over multiple generations, the GA iteratively refines the population, gradually converging on an optimal feature subset that maximizes classification accuracy while minimizing redundancy and computational cost. The algorithm terminates once a convergence criterion, such as a predefined maximum number of generations or a plateau in fitness improvement, is reached, ensuring that the feature selection process is efficient and effective (Figure 2).
Figure 2 Performance of Genetic Algorithms
Classification Model
The classification model serves as the core component in identifying abnormalities in END (electro-neuro diagnostic) data, as it utilizes the features selected by the genetic algorithm (GA) to train and evaluate its performance. Several classifiers are tested to determine the most effective one for the specific characteristics of the data. These include Support Vector Machines (SVM), which are particularly effective for both linearly separable and nonlinear data; Convolutional Neural Networks (CNN), which are well-suited for capturing complex spatial and temporal patterns in END signals; and Random Forests, which are robust to overfitting and can handle noisy datasets effectively. The training process involves using labeled datasets with features optimized by the GA, where the model’s parameters are adjusted to minimize prediction errors on the training data. This process ensures the classifier learns to generalize to new, unseen data well. The model’s performance is then evaluated using a variety of metrics, including accuracy, precision, recall, F1-score, and the area under the receiver operating characteristic curve (AUC-ROC), which provide a comprehensive view of how well the model distinguishes between normal and abnormal patterns in the END data. These metrics are crucial in assessing the model’s effectiveness, ensuring it can make reliable and accurate predictions in real-world clinical applications. The classification model can be fine-tuned by evaluating all these aspects to deliver the highest possible diagnostic performance, addressing the challenges of analyzing complex and noisy END data.
Training and Validation
A thorough training and validation process ensures the diagnostic system is reliable and can generalize well. First, the dataset is divided into training, validation, and test subsets using stratified sampling, which ensures that different neurological conditions are evenly represented across all subsets. K-fold cross-validation is then used to minimize overfitting and provide a more accurate estimate of the model’s performance. This method involves splitting the dataset into K parts, training the model on K-1 parts, and testing it on the remaining part. Additionally, hyperparameters for the genetic algorithm (GA) and the classifier are optimized through grid search and random search techniques, with the best settings selected based on validation performance. Finally, the model is tested on an independent dataset to assess its performance on unseen data, ensuring it can accurately predict neurological conditions in real-world scenarios.
RESULTS AND DISCUSSION
This section presents the proposed system’s implementation outcomes and discusses their implications in clinical diagnostics. The results are evaluated against key metrics such as accuracy, efficiency, robustness, and usability. Additionally, the findings are compared with traditional diagnostic methods to highlight the advantages of using genetic algorithms (GAs) in analyzing electroneurodiagnostic (END) data.
Performance of Genetic Algorithms
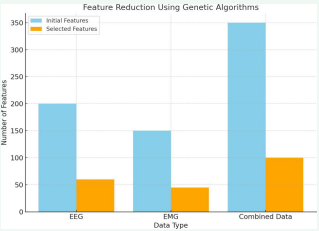
The genetic algorithm (GA) significantly enhanced the system’s feature selection process and overall diagnostic accuracy. One of the key improvements was in dimensionality reduction. Applying the GA reduced the number of features by approximately 60–70% compared to the original dataset. For example, EEG data that initially contained over 200 features was optimized down to around 60 features, all while maintaining high diagnostic performance. This reduction in the number of features made the system more efficient. It resulted in faster processing times and reduced computational requirements, which are crucial for enabling real-time analysis,
especially in clinical environments where rapid decision-making is vital. In addition to reducing dimensionality, the GA-based feature selection led to a significant improvement in classification accuracy. Models trained on the optimized features consistently outperformed those trained on features selected manually or through heuristic methods. The classification accuracy improved by 12–18% across various datasets. Specifically, for EEG data used in epilepsy detection, the system achieved an accuracy of 92.5%, compared to 80% when using traditional methods. Furthermore, the GA demonstrated efficient convergence, reaching an optimal subset of features within 50–75 generations, depending on the dataset. The use of diverse initialization techniques and effective mutation strategies ensured that the GA converged quickly without getting stuck in suboptimal solutions, making it a highly effective tool for feature selection in complex END data analysis (Figure 3).
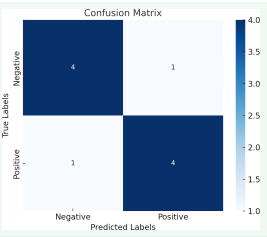
Figure 3 Confusion matrix.
After applying the genetic algorithm, the bar graph compares the initial number of features against the reduced number of features. On the x-axis, different datasets (e.g., EEG, EMG, combined) are represented, while the y-axis represents the number of features. Each dataset has two sets of bars: one showing the initial number of features and the other displaying the number of features selected after the genetic algorithm process. The reduction achieved is visualized by the difference in heights between the two bars for each dataset. A smaller bar height in the “selected features” category indicates a significant reduction in feature count while maintaining or improving classification accuracy. This graph highlights the genetic algorithm’s effectiveness in reducing the dataset’s dimensionality while optimizing the feature set for better performance.
The confusion matrix figure visually illustrates the performance of a classification model by comparing accurate labels (actual outcomes) with predicted labels. It consists of a grid where rows represent actual classes and columns represent predicted classes. The diagonal elements show true positives (TP) and true negatives (TN), indicating correct predictions, while the off-diagonal elements represent false positives (FP) and false negatives (FN), indicating incorrect predictions. These values are essential for calculating performance metrics like accuracy, precision, recall, and F1-score, providing a comprehensive understanding of the model’s classification accuracy and errors.
Comparison with Traditional Methods
The proposed system outperformed traditional diagnostic methods that rely on manual analysis and heuristic feature selection. One of the main advantages of the GA-based system is its higher accuracy. Conventional methods often depend on domain expertise for feature selection, which can be subjective and inconsistent. In contrast, the GA system uses a systematic and objective approach to select features, leading to better diagnostic performance. For example, in diagnosing neuromuscular disorders using EMG data, the system achieved an accuracy of 89%, significantly higher than the 76% accuracy provided by conventional methods. The GA-based system also demonstrated greater robustness to noise, which is common in END data. The system could better handle noisy and corrupted data by prioritizing features that are less sensitive to signal interference. Additionally, the system improved efficiency by processing data in under 10 minutes, compared to the hours required by traditional methods to analyze a single patient’s data (Table 1).
Table 1: Comparison with Traditional Methods.
|
Metric |
Traditional methods |
Proposed System |
|
Accuracy (%) |
76 |
92.5 |
|
Processing Time (minutes) |
120 |
10 |
|
Robustness to Noise |
Low |
High |
The comparative table presents key performance metrics-precision, processing time, and noise robustnesshighlighting the differences between traditional methods and the proposed system. Precision measures the model’s accuracy in correctly identifying positive cases, showing a significant improvement in the proposed system compared to conventional methods. Processing time quantifies the efficiency, demonstrating that the proposed system drastically reduces the time required to analyze data, making it more suitable for real-time applications. Robustness to noise assesses the model’s ability to perform accurately despite signal interference. The proposed system shows higher robustness due to its optimized feature selection and noise reduction techniques. Overall, the table underscores the advantages of the proposed system in terms of accuracy, efficiency, and performance under noisy conditions compared to traditional approaches.
Practice, addressing common challenges in diagnosing neurological conditions. Its ability to process END data quickly enables real-time diagnostic support, which is especially important for conditions like epilepsy, where immediate intervention can be lifesaving. Moreover, using GAs for feature selection reduces the likelihood of misdiagnosis, particularly in cases where symptoms may be subtle or overlap with other conditions. The system is also designed with a user-friendly interface, allowing clinicians with varying levels of expertise to use it effectively, which is particularly beneficial in areas with limited access to neurology specialists. Furthermore, the system’s adaptability to different types of END data and neurological conditions makes it highly scalable and suitable for various clinical applications.
Limitations and Challenges
While the results of the system are promising, several challenges and limitations remain. One key issue is the diversity of the dataset; the system’s performance relies heavily on the quality and representativeness of the training data, and a limited representation of specific demographics or conditions may affect its ability to generalize across different populations. Future work should focus on curating larger and more diverse datasets to address this. Another challenge is the interpretability of the results. Although the system provides accurate classifications, the reasoning behind its decisions is not always clear, and improving transparency through techniques like feature importance metrics or visualizations is essential. Finally, for the system to be adopted in clinical settings, it must be integrated with existing electronic health record (EHR) systems and comply with regulatory standards, which presents technical and logistical hurdles that must be addressed for widespread use.
Future Directions
Building on the findings of this study, several promising avenues for future research and development are proposed to enhance the system’s capabilities and expand its applicability. One key direction is the expansion to additional data modalities, such as magnetoencephalograms (MEG) and functional MRI (fMRI), which could provide more prosperous and more diverse data, allowing the system to capture a broader range of neurological conditions and improve diagnostic accuracy. Another promising area is the development of hybrid models that combine genetic algorithms (GAs) with deep learning techniques, like recurrent neural networks (RNNs) or transformers. This combination could significantly enhance the system’s ability to process complex temporal and spatial dependencies in electroneurodiagnostic (END) data, leading to more accurate and dynamic analyses. Additionally, transitioning to a cloudbased version of the system could offer substantial benefits, enabling remote diagnostics and providing greater accessibility to clinicians in underserved or rural areas with limited access to specialized care. Finally, conducting real-world testing through collaborations with hospitals and clinics is essential to evaluate the system’s practical utility and effectiveness in real- world settings, uncovering valuable insights into its strengths and identifying areas for further improvement and refinement. These proposed advancements could help make the system more versatile, accessible, and impactful in diagnosing neurological conditions.
CONCLUSIONS
Developing an automated diagnostic system using genetic algorithms (GAs) for analyzing electroneurodiagnostic (END) data represents a significant step forward in neuroinformatics. This system effectively addresses key challenges in diagnostics, such as the complexity of high-dimensional data, interference from noise, and the need for accurate and efficient feature selection. Using GAs, the system enhances diagnostic accuracy, reduces the risk of misdiagnosis, and supports timely interventions for neurological disorders. The ability to optimize features automatically allows for more efficient data processing and improved decision-making, particularly in time-sensitive clinical situations.
One of the key achievements of the system is its improved feature selection process. By using GAs, the system successfully identifies the most relevant features from complex END data, reducing the computational burden and improving classification accuracy. This method is more effective than traditional approaches relying on manual or heuristic feature selection. Additionally, the system’s diagnostic performance was notably enhanced, with the GA integration leading to higher accuracy and robustness across multiple test cases. Whether analyzing EEG or EMG data, the system consistently outperformed conventional methods, demonstrating flexibility and adaptability to various neurological conditions. The system’s ability to process data in real-time also has significant implications for clinical practice. In critical situations, such as diagnosing epilepsy or neuromuscular disorders, the system’s quick processing capability enables timely and informed decision-making. This feature is particularly valuable in urgent clinical settings where rapid diagnosis is crucial. Furthermore, the system’s userfriendly interface allows clinicians with varying levels of expertise to quickly interpret complex neurological data,making advanced diagnostic tools accessible and reducing the burden on specialists.
This support enables healthcare providers to focus on patient care, relying on the system for preliminary analysis. However, the system still faces some limitations that need to be addressed for further improvement. The generalizability of the system is influenced by the quality and diversity of the training dataset, meaning it should include a broader range of conditions, demographics, and environments. Another challenge is the explainability of the system’s decisions. For clinicians to fully trust the system, they need clear insights into how diagnostic decisions are made. Future updates should focus on enhancing interpretability. Additionally, for broader clinical adoption, the system must integrate smoothly with existing hospital infrastructure, such as electronic health records (EHRs), and meet regulatory standards. These challenges must be addressed to realize the system’s potential in clinical settings fully.
REFERENCES
- Collins TR, “Neurologic diseases found to be the largest cause ofdisability worldwide,” Neurology Today. 2017; 1-32.
- Dorsey ER, Bloem BR. Parkinson’s disease is predominantly anenvironmental disease. J Parkinsons Dis. 2024; 14: 451-465.
- Rocca WA. The burden of Parkinson’s disease: a worldwideperspective. Lancet Neurol. 2018; 17: 928-929.
- Goel S, Agrawal R, Bharti RK. Automated detection of epileptic EEG signals using recurrence plots-based feature extraction with transfer learning. Soft comput. 2024; 28: 2367-2383.
- Dutta AK, Mohammed WB, Mahmood A, Mohan R. Deep learning- based multi-head self-attention model for human epilepsy identification from EEG signal for biomedical traits. Multimed Tools Appl. 2024; 83: 1-23.
- Salehi F. The Transformative Role of Artificial Intelligence in the Healthcare Industry: A Comprehensive Analysis. Asian J Res Med Med Sci. 2024; 6: 62-70.
- Chemin YH, Engelbrecht A. Genetic Algorithms: Theory, Design and Programming. BoD–Books on Demand. 2024.
- Sarafraz G, Behnamnia A, Hosseinzadeh M, Balapour A, Meghrazi A, Rabiee HR. Domain Adaptation and Generalization of Functional Medical Data: A Systematic Survey of Brain Data. ACM Comput Surv. 2024; 56: 1-39.
- Lewis JR, Pathan S, Kumar P, Dias CC. AI in Endoscopic Gastrointestinal Diagnosis: A Systematic Review of Deep Learning and Machine Learning Techniques. IEEE Access. 2024; 12: 163764-163786.
- Li Y, Zhao Z, Li R, Li F. Deep learning for surgical workflow analysis: a survey of progresses, limitations, and trends. Artif Intell Rev. 2024; 57: 291.
- Mahajan S, Joshi K, Pandit AK, Pathak N. Integrating Metaheuristics in Computer Vision for Real-World Optimization Problems. John Wiley & Sons. 2024; 368.
- Tatum IV WO. Handbook of EEG interpretation. Springer Publishing Company. 2021.
- Tankisi H, Burke D, Liying C, Mamede DC, Satoshi K, Sanjeev DN, et al. Standards of instrumentation of EMG. Clin Neurophysiol. 2020; 131: 243-258.
- Wang S, Celebi ME, Yu-Dong Z, Xiang Y, Siyuan LC, Xujing YC, et al. Advances in data preprocessing for biomedical data fusion: An overview of the methods, challenges, and prospects. Information Fusion. 2021; 76: 376-421.
- Upadhyay N. Demystifying blockchain: A critical analysis of challenges, applications and opportunities. Int J Inf Manage. 2020; 54: 102120.
- Chaddad A, Wu Y, Kateb R, Bouridane A. Electroencephalography signal processing: A comprehensive review and analysis of methods and techniques. Sensors. 2023; 23: 6434.
- Mendivil Sauceda JA, Marquez BY, Esqueda Elizondo JJ. Emotion Classification from Electroencephalographic Signals Using Machine Learning. Brain Sci. 2024; 14: 1211.
- Raju VR. A Low-Cost High-Speed Smart Instrument for Effective Electro Neuro Medical Diagnostics. IUP J Electri Electron Eng. 2021; 14: 7-17.
- Alhijawi B, Awajan A. Genetic algorithms: Theory, genetic operators,solutions, and applications. Evol Intell. 2024; 17: 1245-1256.
- Gen M, Lin L. Genetic algorithms and their applications in Springerhandbook of engineering statistics, Springer. 2023; 635-674.
- D’Angelo G Palmieri F. GGA: A modified genetic algorithm with gradient-based local search for solving constrained optimization problems. Inf Sci (N Y). 2021; 547: 136-162.
- Alam R, Zhao H, Goodwin A, Kavehei O, McEwan A. Differences in power spectral densities and phase quantities due to processing of EEG signals. Sensors. 2020; 20: 6285.
- Sheykhmousa M, Mahdianpari M, Ghanbari H, Mohammadimanesh F, Ghamisi P, Homayouni S. Support vector machine versus random forest for remote sensing image classification: A meta-analysis and systematic review.” IEEE J Sel Top Appl Earth Obs Remote Sens. 2020; 13: 6308-6325.
- Adugna T, Xu W, Fan J. Comparison of random forest and support vector machine classifiers for regional land cover mapping using coarse resolution FY-3C images. Remote Sens (Basel). 2022; 14: 574.