“Everything Out” Validation Approach for Qsar Models of Chemical Mixtures
- 1. Department of Molecular Structure and Cheminformatics, AV Bogatsky Physical Chemical Institute, Ukraine
- 2. Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, University of North Carolina, USA
- 3. Department of Chemical-Technological, Odessa National Polytechnic University, Ukraine
- 4. Institute of Computer Systems, Odessa National Polytechnic University, Ukraine
Abstract
Established strategies for validating QSAR models of binary mixtures of chemicals are not applicable to the most challenging case, which is the prediction of binary mixtures created by two compounds not present in the initial training set. In this study, we have addressed this challenge by introducing the “Everything Out” validation strategy where the external sets are deliberately formed by all binary combinations of two compounds excluded from the training set. The model accuracy is evaluated by the error of prediction for the external sets. We show that the “Everything Out” approach affords lower error of prediction for binary mixtures formed by two new compounds and similar error of prediction for mixtures with one new compound as compared to the alternative “Compound Out” validation approach. We posit that “Everything Out” should be employed as the preferred approach to validating QSAR models of binary mixtures.
Keywords
• External validation
• Molecular modeling
• Structure-activity relationship
• QSAR of mixtures
Citation
Muratov EN, Varlamova EV, Kuzmin VE, Artemenko AG, Muratov NN, et al. (2014) “Everything Out” Validation Approach for Qsar Models of Chemical Mixtures. J Clin Pharm 1(1): 1005.
INTRODUCTION
Chemical mixtures are widely used in products of pharmaceutical industry, agriculture, and cosmetics. The experimental safety testing of individual environmental organic compounds presents significant challenges as illustrated by extensive and costly projects such as REACH [1] or TOX21 [2]. These experimental challenges are dramatically exacerbated in the case of chemical mixtures due to the complexity of their compositions including molecular diversity and the relative ratios of their components. Nevertheless, since manufactured compounds rarely enter the environment independently, the evaluation of potential health and safety impacts of compound mixtures is perhaps even more critical than that of individual chemicals.
Computational approaches such as cheminformatics, especially QSAR modeling, may provide an effective alternative to experimental methods to reduce time and cost of developing new mixture formulations with the desired properties and safety profiles [3]. However, although modern QSAR methodology is fairly successful in dealing with individual compounds, there are no mature, well-established approaches that could be directly used to model properties of mixtures. This is mostly due to the absence (or lack) of reliable experimental data on mixture properties, adequate descriptors of mixtures, and robust strategies for the external validation of developed models. To the best of our knowledge, the issue of rigorous QSAR modeling of mixtures has been addressed only in a few publications [3–5].
Rigorous external validation is the integral part of any QSAR exercise, irrespective to the nature of the chemical objects under investigation [5]. However, proper external validation of QSAR models for mixtures is much less straightforward in comparison to traditional QSAR analysis [3]. Here, the conventional external cross-validation procedure,[6] when individual compounds are randomly placed in the external set (or fold), i.e., no information regarding excluded compound is present in the training set, is not scrupulous enough. The reason is obviously due to the fact that in traditional QSAR application each entry in a dataset is a single compound whereas a mixture consists of at least two compounds that could be blended in different ratios, i.e., each mixture could be represented by several entries. In traditional QSAR, the placement of several randomly-selected compounds into the external set will result in the complete absence of structural information about these compounds in the training set. On the contrary, in QSAR modeling of mixtures the information about a mixture created by certain compounds would be still available because other entries corresponding to the same mixture with different ratios of the same components will remain in the training set. As a result, the model’s predictive performance will be over-estimated. These considerations prompted us to start devising more rigorous protocols for external validation of QSAR models of mixtures [4,5].
Previously, we have introduced three different strategies for external validation depending on the initial data and the actual application of developed models:[5] (i) “Points Out” – prediction of the investigated property for any composition of the mixtures from modeling set, (ii) “Mixtures Out” –filling of missing data in the initial mixtures’ data matrix (i.e., prediction of the investigated property for mixtures with unknown activity created by pure compounds from the modeling set), and (iii) “Compounds Out” – prediction of the investigated property for mixtures formed by a novel pure compound that was absent in the modeling set. These strategies address the situations of predicting new mixtures created by (i) two compounds from the modeling set and (ii) a new compound and a compound from the existing matrix of mixtures. However, the most interesting and the most difficult case of evaluating the model accuracy for predicting a mixture created by two new compounds still remains uncovered.
The goal of this study is to introduce the “Everything Out” validation strategy for QSAR modeling of mixtures. This procedure simulates the addition of novel compounds to the existing matrix of mixtures and gives a reasonable idea about the expected error of prediction for the mixtures created by two new compounds that were absent in the modeling set. Although the error of prediction for this strategy is expected to be the largest, QSAR models passing “Everything Out” validation should be able to predict the investigated property for mixtures created by the compounds outside of the modeling set taking into account models’ applicability domain. Thus, we posit that “Everything Out” is the most rigorous method for external validation of QSAR models of mixtures.
MATERIALS AND METHODS
“Everything Out” validation strategy
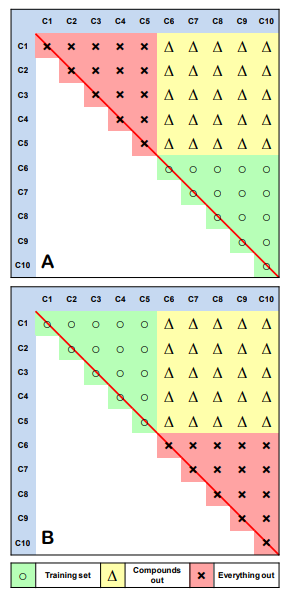
Following this new strategy, the data matrix of mixtures is divided into three parts (see Figure 1A).
Figure 1: "Everything Out" strategy of external validation in QSAR modeling of mixtures.
For example, let’s consider completely filled matrix of mixtures created by 10 compounds. The first part (compounds C1-C5 and all of their binary mixtures) is used as a training set; the second part (compounds C6-C10 and all of their mutual mixtures) is used as the “Everything out” external set; and the remaining part is employed as “Compounds out” external set [5]. It is important to stress that mixtures created by compounds from the same group belong only to either “training” or “everything out” part and not to both of them simultaneously; meanwhile, mixtures created by compounds from different parts create the “compounds out” part (see Figure 1). Then “training” and “everything out” sections are switched, i.e., “training” part becomes the “everything out” one and vice versa (Figure 1B). “Compounds out” part remains the same for both folds. Thus, every mixture in the “everything out” set is always created by two compounds that are absent in the training set. If the mixture matrix is completely filled, compounds that created this matrix could be sorted randomly or alphabetically. In case of a sparse data matrix, supervised selection of training and test sets is needed to keep the size of the sets more or less equal. However, even despite the supervised process of fold creation, sometimes “everything out” and “compound out” folds could be predicted poorly because some of them can be created mostly by compounds and mixtures that are very different from those in the training set. One could shuffle (re-order) the matrix of mixtures several times and repeat modeling to obtain more consistent prediction performances.
Data set
There is still a significant lack of experimental data for mixtures. Therefore, for the purposes of this study we have used the vapor/liquid equilibrium diagrams for bubble point temperatures of binary liquid mixtures we modeled earlier [4]. The dataset consisted of 67 pure liquids and 167 mixtures of these liquids. Each mixture was represented by several (7-57) points; thus, 167 mixtures in the modeling set have been described by 3,185 data points. More details about this dataset could be found elsewhere [4].
RESULTS AND DISCUSSION
The models were built using random forest and SiRMS descriptors [7] and validated using three strategies described above, i.e., “Points Out”, “Mixtures Out”, and “Compounds Out”. Detailed description of model building and validation could be found elsewhere [4]. Then, an independent external set consisting of 94 new mixtures made of 66 compounds was used for model validation as well. Among these 94 mixtures (632 data points), 27 combinations contain no new pure compounds, 63 mixtures (1,386 data points) contain one new compound, and four remaining mixtures were created by compounds that were absent in the modeling set. The results of 5-fold external cross-validation and performance of the models obtained using “Compounds Out” and “Everything Out” strategies are shown in Table 1. External set mixtures containing one and two new compounds were treated separately in order to estimate the error for the corresponding validation strategy (“Compounds Out” and “Everything Out” respectively).
We have preserved the initial splitting for modeling and external sets, model building and validation workflow, and the applicability domain estimation procedure from the previous study [4]. “Everything Out” set was formed from the modeling set compounds as shown in Figure 1. Twenty eight splits were generated in order to achieve more consistent results and to insure that every mixture was present in the “Everything Out” set at least once. Then, developed models were applied to the external set consisted of 95 mixtures (2065 data points). As obvious from the results (see Table 1), the RMSE for the “Compounds out” set is comparable to that obtained in the previous study,[4] i.e., 12.1 K vs 10.3 K. It means that, using “Everything Out” strategy, we could adequately estimate the error for mixtures containing one new component. However, the RMSE for “Compounds Out” estimated on the external set is significantly higher (~19K).
Expectedly, the error of prediction for “Everything Out” strategy estimated on the modeling set using 5-fold external cross-validation is higher than that for “Compounds out” strategy (17.1 K vs 12.1 K). This is fully in tune with our expectations that it is harder to predict a mixture containing two new components than a mixture with one new component. Meanwhile, the results obtained on the external set are not as encouraging because the error of prediction is somewhat higher (~23 K). However, we have to emphasize that the “Everything Out” set was very small (only eight compounds creating four sets of mixtures), and after taking into account the applicability domain for filtering out chemicals too dissimilar from the modeling set compounds, it was reduced to only four compounds and two sets of mixtures (44 data points). Thus, one must be extremely cautious with RMSE values computed for this very limited number of mixtures. Certainly, the new data obtained with a larger set of mixtures created by two new compounds are needed to make this comparison more reliable. However, our results clearly show that (i) “Everything out” strategy has similar performance with “compounds out” strategy for estimating the prediction error for the mixtures including one new compounds absent in a modeling set; (ii) “Everything Out” is more rigorous and thus more suitable for estimating the prediction error for mixtures created by both compounds absent in a modeling set than the “Compounds Out” strategy.
Table 1: RMSE (K) for different strategies of external QSAR model validation.
| Modeling set, 5-FECVa | External set | |||
| 1 new compound | 2 new compounds | 1 new compoundb | 2 new compoundsc | |
| Compounds Out[4] | 10.3 | NA | 18.8 | 23.1 |
| Everything Out | 12.1 | 17.1 | ||
Abbreviations: a 5-fold external cross-validation; b 62 combinations containing one new compound absent in the modeling set;
c 2 combinations containing two new compounds absent in the modeling set.
CONCLUSION
In conclusion, we have developed a robust and useful modeling and validation protocol to predict the properties of binary mixtures created by new compounds not found in the modeling set. This approach is universal and could be used for assessing the prediction error for both binary mixtures containing just one new component (expanding upon the application of “Compounds Out” strategy developed by us earlier) as well as for mixtures created by two new compounds. We suggest that the “Everything Out” strategy should be used as the method of choice in developing and validating QSAR models of mixtures.
ACKNOWLEDGEMENT
E.M., D.F., and A.T. gratefully acknowledge the financial support from NIH (grant GM66940) and EPA (RD 83382501 and R832720). E.M., A.A., and V.K. are thankful to STCU (Project 407) for the financial support. A.T. acknowledges partial support from Russian Scientific Foundation (project 14-43-00024).
REFERENCES
- European Comission. REACH: Registration, Evaluation and Authorisation and Restriction of Chemicals.
- US EPA. Tox21. 2014.
- Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, et al. QSAR Modeling: Where have you been? Where are you going to? J. Med. Chem. 2014; 57: 4977–5010.
- Oprisiu I, Varlamova E, Muratov E, Artemenko A, Marcou G, Polishchuk P, et al. QSPR Approach to Predict Nonadditive Properties of Mixtures. Application to Bubble Point Temperatures of Binary Mixtures of Liquids. Mol. Inform. 2012; 31: 491–502.
- Muratov EN, Varlamova E V., Artemenko AG, Polishchuk PG, Kuz’min VE. Existing and Developing Approaches for QSAR Analysis of Mixtures. Mol. Inform. 2012; 31: 202–221.
- Tropsha A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010; 29: 476–488.
- Kuz’min VE, Muratov EN, Artemenko AG, Varlamova EV, Gorb L, Wang J, et al. Consensus QSAR Modeling of Phosphor-Containing Chiral AChE Inhibitors. QSAR Comb. Sci. 2009; 28: 664–677.