Identification and Functional Characterization of an Alternative 5
- 1. University of Manitoba, Department of Human Nutritional Sciences, University of Manitoba, Winnipeg, Canada
- 2. Molecular and Clinical Nutrition Section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA
Abstract
Since the initial description of the human SLC23A1 transcript, encoding a sodiumdependent ascorbic acid transporter, no alternative splice variant had been described. In this report we describe a novel human specific alternative first exon which is located 1078 nucleotides 5’ of the canonical first exon. The two first exons are not mutually exclusive since they splice together to create a novel SLC23A1 protein isoform, we name it isoform 1A, with a N-terminus that is elongated by 36 amino acid. The novel SLC23A1 isoform locates to the plasma membrane, but mediates only 7% of the ascorbic acid transport exhibited by the shorter isoform when expressed in Xenopus laevis oocytes.
Keywords
• SLC23A1
• Alternative first exon
• Ascorbic acid transport. SLC23A1
• Alternative first exon
• Ascorbic acid transport
Citation
Shaghaghi MA, Tu H, Yurkova N, Levine M, Eck P (2013) Identification and Functional Characterization of an Alternative 5’ Exon of the Sodium Dependent Ascorbic Acid Transporter SLC23A1. J Hum Nutr Food Sci 1: 1001.
ABBREVIATIONS
SLC23A1- Solute Carrier 23 A1, also known as sodiumdependent ascorbic acid membrane transporters 1; alternative name: SVCT1; HEK- Human Embryonal Kidney cells; PKCProtein Kinase C (PKC) (phosphorylation sites); tGFP- turbo Green Fluorescent Protein; RFP- Red Fluorescent Protein; ESTExpressed Sequence Tags; BLAST- Basic Local Alignment Search Tool; RNA- Ribonucleic Acid; cRNA-complementary Ribonucleic Acid; VISTA- Visualizing Global DNA Sequence Alignments of Arbitrary Length; PCR- Polymerase Chain Reaction; pI- Isoelectric point (pH at which a particular molecule or surface carries no net electrical charge.); ORF-Open Reading Frame; kDa- Kilo Dalton (a permitted non-SI unit for molecular mass or mass of a particular band in a separating gel); M- Molar.
INTRODUCTION
The SLC23A1 locus on human chromosome 5q31.2 (138702885-138719039 compl; NC_000005.9) encodes for the sodium-dependent ascorbic acid transporter 1. Ascorbic acid, the bioactive form of Vitamin C is essential for human survival and the prevention of common complex diseases [1,2]. Main sites of SLC23A1 expression are the small intestine, liver and kidney, where it functions as an apical ascorbic acid uptake carrier [3- 9]. SLC23A1 transports ascorbic acid with moderate affinity (Km ~ 100-200 µM) and high capacity, reflecting its role in intestinal absorption and renal re-absorption [3,4,10]. Two alternative splice variants have been described in the literature and are listed in the NCBI database as reference RNAs NM005847.4 and NM152685.3. The protein isoform represented by the splice variant NM152685.3 mediates cellular ascorbic acid uptake [11] [5,12], while the isoform encoded by NM005847.4, which contains additional 12 nucleotides between exon 5 and 6 adding 4 amino acids, shows no ascorbic acid transport [5].
The SLC23A1 exon intron structure has been described as containing 15 exons, stretching over 16 kilobases [13,14], and is similar to the mouse ortholog [15]. SLC23A1 is the key protein in renal re-absorption and hepatic maintenance of ascorbic acid [16], making it a major determinant of systemic ascorbic acid levels. Genetic variation in SLC23A1 has a high potential to impact on systemic ascorbic acid levels, and the potential detrimental health impact is demonstrated by the fact that the slc23a1-/- mouse has low plasma and tissue concentrations and ~50% perinatal mortality [16].
Most genes in the human genome have more than two splice variants [22]; therefore we evaluated the existence of additional SLC23A1 transcripts in silico. In this report we characterize a novel alternative first exon encoding a SLC23A1 isoform which is elongated by 36 additional N-terminal amino acids on a genomic and functional level.
MATERIAL AND METHODS
Expressed sequence tags (EST) search and alignments
All mRNA and EST sequences in the human SLC23A1 5’ region were identified by BLASTing the known SLC23A1 Exon (1B) and intron 1, as well as the 5000 nucleotides in the proximal promoter region (NCBI Human Genome Build 37.3) against the human Non-RefSeq RNA and EST databases (http://blast.ncbi. nlm.nih.gov/Blast.cgi). Data were downloaded, assembled and visually analyzed (curated) using Sequencher 4.9, changing alignment parameter as needed. Data were curated and validated visually and compared to data available from the NCBI and Ensembl databases for verification. Evidence for expression in non humanoid life forms was evaluated through BLAST searches in databases for all available species (http://blast.ncbi.nlm.nih. gov/Blast.cgi).
Comparative conservation analysis using VISTA tools
Using the human genome (NCBI Human Genome Build 37.3) as the base genome, candidate orthologous regions were identified using gVISTA (http://genome.lbl.gov/cgi-bin/ GenomeVista). The sequence of the novel SLC23A1 Exon1A was blasted in gVISTA (http://genome.lbl.gov/cgi-bin/GenomeVista) to pinpoint its precise genomic location. Conservation in the latest pre-computed genomic assemblies were compared in the VISTA browser (http://pipeline.lbl.gov/cgi-bin/gateway2?select or=vistapoint).
Real Time PCR
Real Time PCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) following manufactures recommendations. StepOnePlus operation was controlled by the StepOne software vs. 2.2.2, which also used for data processing and analysis. Final analysis was confirmed using Microsoft Excel, which was also used to design graphics.
PrimeTime® TaqMan based qPCR assay IDT47803527 (Integrated DNA Technologies) was used to determine levels of the canonical consensus SLC23A1 transcript represented by NM005847.4 and NM152685.3. PrimeTime® TaqMan based qPCR assay IDT52386812 (Integrated DNA Technologies) was used to determine levels of the transcript containing the novel SLC23A1 exon 1A. Assays were performed in TaqMan Fast Advanced Master Mix (Applied Biosystems/Invitrogen) following the manufacturers protocols. Clontech human liver, small intestine and kidney Marathon ready cDNA (Clontech), which had been standardized based on β–actin levels by the manufacturer, was used in the qPCR. Plasmids representing the SLC23A1 consensus as well as exon 1A transcript were used in serial dilutions to determine a standard curve, where plasmids copy numbers were calculated based on the formula: m=(n)x1.096e-21g (m=mass, n=plasmid size in nucleotides).
Cloning of the SLC23A1 exon1A cDNA
For all PCR based subcloning steps Phusion High-Fidelity DNA Polymerase (New England BioLabs) was used according to the manufacturers recommendations. The identity of PCR amplicons and the final inserts in the DONR and expression plasmids had been confirmed by sequencing (The Centre for Applied Genomics, TCAG) and sequence data were analyzed by Sequencher 5.0 (GeneCodes). A two step PCR methodology was applied to create the fluorescently tagged protein, where first two separate PCR products for the SLC23A1 exon 1A ORF (template RNA was obtained from CaCo-2 cells) and turbo Green Fluorescent Protein (tGFP) (template GIPZ Lentiviral shRNAmir vector, Open Biosystems) were produced (Table of primers in Supplement). These amplicons were designed to have overlapping sequences and in a second round of PCR these were used to obtain one amplicon, which was tagged with the GatewayTM (Invitrogen) recombination tags. This amplicons was integrated into pDONR22.1 and transferred into pcDNA/V5- DEST using the manufacturer’s protocol. The ORF representing the shorter SLC23A1 splice variant (NM005847.4) was contained in the m-cherry red fluorescent protein vector mCherry-SLC23A1 pReceiver-M55 (GeneCopoeia).
Cell Culture and Transfection
HEK293 cells (ATCC) were cultured at 37°C in DMEM/F-12 HyClone media (Thermo Fisher Scientific), 10% fetal bovine serum (FBS) on 75mm flasks (Thermo Fisher Scientific, Corning) or 24-well tissue culture plates (Thermo Fisher Scientific, BioLite). At appropriate densities the cells were transfected with mCherry-SLC23A1 pReceiver-M55 or tGFP-SLC23A1 Exon1A pcDNA/V5-DEST using the Effectene Transfection Reagent (Quiagen) following the manufacturer’s protocols. 24 to 48h post transfection the cells were visualized by fluorescent microscopy (Axiovert 200, ZEISS).
Fluorescent Microscopy
24 to 48h post transfection the fluorescence in the transfected cells was visualized using an Axiovert 200M inverted wide-field epifluorescence microscopy (Carl Zeiss). Images were collected with the Axiovision software package AxioVS40 v 4.5.
Protein prediction analysis
ProtParam (http://web.expasy.org/cgi-bin/protparam/ protparam) was used for computation of various physical and chemical parameters, including the molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity (GRAVY). The TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict the transmembrane architecture of the proteins. PredictProtein (https://www.predictprotein.org) was used to predict secondary structure, solvent accessibility, transmembrane helices, globular regions, coiled-coil regions, structural switch regions, B-values, disorder regions, intraresidue contacts, protein-protein and protein-DNA binding sites, sub-cellular localization, domain boundaries, beta-barrels, cysteine bonds, metal binding sites and disulphide bridges of the two proteins.
Xenopus laevis Oocytes Transport Assay
Complementary RNAs (cRNA) for both human SLC23A1 splice variants were prepared by in vitro transcription using the SP6 mMessage mMachine Kit (Ambion). The cRNAs were injected into Xenopus laevis oocytes and the transporter experiments were performed exactly as described [15].
RESULTS
A novel alternative first SLC23A1 exon exclusive to humans is expressed in the small intestine
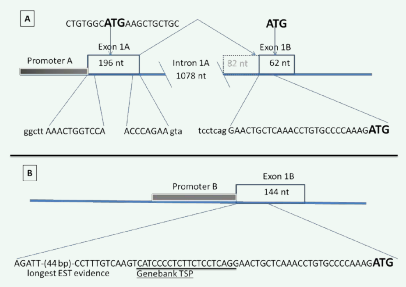
The existence of an alternative first SLC23A1 exon is supported by five Expressed Sequence Tags (ESTs: DA420756, DB480870, DA870503, EG327910, EG328033) aligning 1078 nucleotides 5’ of the currently known SLC23A1 exon 1 (Figure 1A). These five ESTs define a novel first exon for the human SLC23A1 gene, which we name SLC23A1 exon 1A (Figure 1A). The novel SLC23A1 exon1A consists of 196 nucleotides, with the transcript splicing into the following exon sixty-two nucleotides from its 3’ end (Figure 1B). The full length of the exon was confirmed by PCR amplification using RNA from the intestinal carcinoma cell line CaCo-2 as template and subsequent sequence analysis.
Figure 1: Exon usage in the 5’ region of the SLC23A1 gene.
A) Two alternative first exons are utilized. Exon 1A is the novel exon described in this report. Both first exons have independent promoter regions, but are not mutually exclusive, since exon 1A splices into exon 1B, which is the exon described previously [13,14] and defined by the reference RNAs NM005847.4 and NM152685.3.
B) The Transcriptional Start Point (TSP) of SLC23A1 exon 1B is not represented by the reference RNAs; instead it extends 61 nucleotides 5’, which brings the size of exon 1B to 144 nucleotides.
The novel SLC23A1 exon1A is not reflected through any reference RNA in GeneBank. The reference RNAs NM005847.4 and NM152685.3 define the currently documented first exon [13,14] which by virtue of chromosomal order is defined as SLC23A1 Exon 1B (Figure 1B). Find a detailed description of this exon in a separate paragraph below.
The expression of the novel SLC23A1 exon 1A is limited to humans, no transcript with a sequence homology of 70% or higher could be identified in any RNA or EST database. An analysis of the genomic region surrounding SLC23A1 Exon 1A shows conservation in primates, moderate conservation between human and equine (horse), canine (dog), and bovine (cow), and very low conservation between primates and rodents (mouse and rat) (Figure 2).
Figure 2: Comparative VISTA Analysis of the evolutionary conservation of the orthologous regions containing the novel SLC23A1 Exon 1A and the previously characterized SLC23A1 Exon 1B. SLC23A1 is located on the complement strand of chromosome 5. The first lane shows the exact location of the novel SLC23A1 Exon 1A, as determined in a gVISTA blast search. The threshold for high conservation was set to be 70%. SLC23A1 Exon 1A is not conserved in rodents, and moderately conserved in dog and cow, while SLC23A1 Exon 1B is highly conserved across all species.
The low evolutionary conservation, specifically in the proximal promoter region, supports the evidence that this splice variant appears to be unique to humans or primates.
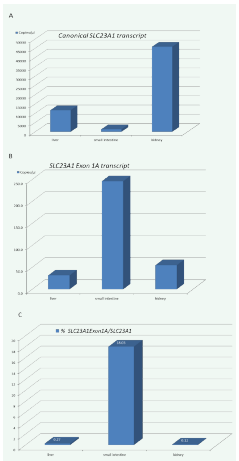
Real time PCR analysis shows abundant expression of the full length SLC23A1 transcript in the kidney, liver and small intestine (Figure 3A), as previously observed [5,11,12,16,17]. In contrast, transcripts containing SLC23A1 exon 1A are only detectable in trace amounts in liver and kidney (Figure 3B), but constitute 18% of the SLC23A1 transcripts in the small intestine (Figure 3C).
Figure 3: Expression of differentially spliced human SLC23A1 transcripts in liver, small intestine, and kidney.
A) The consensus transcript is highly abundant in all tissues confirming prior observations [5,11,12,16,17].
B) Exon 1A is only detectable in trace amounts in liver and kidney, but moderately expressed in the small intestine. C) 18.3% of transcripts in the small intestine contain exon 1A, while less than 03% of transcripts in liver and kidney contain exon 1A.
The expression pattern strongly indicates a tissue specific utilization of exon 1A, with significant abundance in the small intestine. The novel SLC23A1 exon 1A transcript has a total length of 2473 nucleotides (sequence in the appendix).
The novel SLC23A1 protein isoform adds 36 N-terminal amino acids but does not alter transmembrane topology or intracellular location
The novel SLC23A1 Exon 1A contains a translation initiation codon (ATG) (Figure 1A), defining the start of a 1905 nucleotide Open Reading Frame (ORF) (Sequence in the supplement). The resulting novel 634 amino acids containing protein, we name it SLC23A1 isoform A, is in frame with the previously described 598 amino acid containing SLC23A1 protein (NP689898 [5- 11]), adding 36 additional N-terminal amino acids (Supplement Figure 1, and protein sequences).
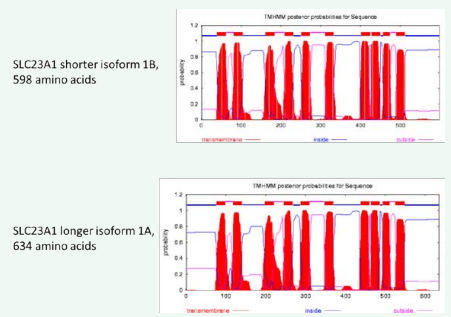
The novel SLC23A1 isoform 1A containing 634 amino acids have a predicted molecular weight of 68.9 kDa. The additional 36 N-terminal amino acid are predicted to change the ratio of negatively charged to positively charged amino acids from 41/37 to 41/44, respectively, changing the charge of the protein from -4 to +3. The theoretical pI is changed from 6.16 for the short isoform B (NP689898) to 8.09 for the novel longer isoform A. Both proteins are predicted to be stable and aliphatic index and grand average of hydropathicity are similar. Furthermore, no changes in membrane topology are predicted, with the N- and the C-termini predicted to be cytosolic (Figure 4).
Figure 4: Predicted membrane topology for the SLC23A1 protein isoforms.
A) The reference protein NP689898 represents the shorter SLC23A1 isoform 1B, and 10 transmembrane domains are predicted.
B) The additional 36 nucleotides in the novel SLC23A1 isoform 1A are not predicted to change the membrane topology, but add to the length of the N-terminal region.
Three novel protein-protein binding sites are predicted in the novel N-terminus of SLC23A1 protein isoform 1A (Supplement Figure 2), and two additional protein kinase C phosphorylation sites are predicted, indicating an altered activation pattern for this isoform (Appendix Figure 2). No signal peptide sequences are identified in both isoform, indicating similar sub-cellular location.
The novel SLC23A1 protein isoform 1A locates to the plasma membrane
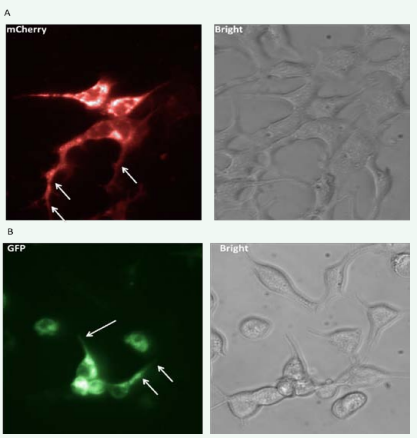
When transiently expressed in human embryonic kidney cells (HEK 293), the shorter SLC23A1 isoform B locates on the outer plasma membrane (Figure 5A), as has been described before [9,18]. The longer SLC23A1 isoform A shows the same membrane localization (Figure 5B). Both isoforms exhibit strong expression when they are solely expressed in the HEK293 cells.
Figure 5: A) mCherry (red) tagged SLC23A1 isoform 1B locates to the plasma membrane in HEK 293 cells.
B) tGFP (green) tagged SLC23A1 isoform 1A also locates to the plasma membrane in HEK 293 cells.
The novel SLC23A1 protein isoform 1A mediates very low ascorbic acid transport in Xenopus laevis oocytes
When expressed in Xenopus laevis oocytes the novel SLC23A1 isoform 1A mediated about 7% of the transport capacity of isoform B (Figure 6).
Figure 6: The novel SLC23A1 isoform 1A mediates ascorbic acid uptake in Xenopus laevis oocytes. [14C]ascorbic acid uptake into Xenopus laevis oocytes injected with cRNA encoding SLC23A1 isoform 1A [Δ] is elevated compared to sham injected oocytes [◊], however, it is only about 7% compared to SLC23A1 isoform 1B [?], when incubated with 300 μM [14C] ascorbic acid.
Definition of the transcriptional start point of SLC23A1 Exon 1B
EST alignments in the 5’ region of SLC23A1 exon 1B shows multiple transcripts extending beyond the previously described transcriptional start point defined by reference RNAs NM005847.4 and NM152685.3 (Figure 1B) [13,14]. PCR amplification of RNA from human intestinal tissue confirm a 144 nucleotides spanning exon 1B, including a 108 nucleotides 5’ untranslated region (5’UTR) (Figure 1B). The total size of the transcript of the shorter SLC23A1 splice variant 1B is therefore 2355 nucleotides from TSP to poly-A signal (sequence in the supplement).
DISCUSSION
This report describes a previously uncharacterized alternative first exon of the SLC23A1 gene, called SLC23A1 exon 1A, which is located 1078 nucleotides 5’ of the currently known SLC23A1 exon 1B and is preferably expressed in the small intestine. It encodes a protein isoform that contains additional 36 N-terminal amino acids, locates to the outer cell membrane, but mediates only about 7% of the ascorbic acid transport of the shorter isoform. The reason for this lack of transport activity is currently not apparent, since both isoforms are predicted to have the same membrane architecture and the molecular imaging confirmed that both locate to the outer cell membrane in mammalian cells. However, the lack of transport activity might be explained by two mechanisms. First, the net charges of the two proteins are reversed, from -4 to +3 and the pI is significantly altered from 6.16 to 8.09 between isoform 1B and isoform 1A, respectively. Therefore simple structural alteration might cause the decreased function. Secondly, protein activation could be modified by two additional protein kinase C (PKC) phosphorylation sites and three novel protein-protein binding sites in the additional N-terminus. PKC phosphorylation had been shown to inhibit membrane proteins activity through decreased structural flexibility as well as modifying the effects of other protein kinases such as protein kinase G and protein kinase A [19]. In addition, PKC activity had been shown to downregulate SLC23A1 via derecruitment from the plasma membrane [20,21].
The existence of the less functional SLC23A1 isoform A might indicate redundancy, specifically since the locus is not evolutionary conserved and the transcript for SLC23A1 variant A is only found in humans. The simple lack of strong evolutionary pressures to decrease the size of the human transcriptome might explain the existence of a less functional membrane transporter isoform [22]. However, this theory might not apply to a human intestinal ascorbic acid transporter, since primates lost the ability to synthesize ascorbic acid and solely rely on intestinal absorption. Therefore the recently developed concept that transporter efficiency is fine-tuned to specific ranges of substrate concentration offers an explanation for the existence of SLC23A1 isoform 1A [22]. Since ascorbic acid concentrations in the small intestine vary greatly, the amino acid composition of the different isoforms might be adapted to provide optimal substrate/transporter interactions. Substrate concentrations in the small intestine can exceed 300 µM, which was tested in the presented experiments, and isoform A might have better efficiency in higher substrate concentrations, where isoform B reaches maximal saturation [1,4-6]. This would also explain the lack of SLC23A1 isoform A in the kidney and liver, where maximal ascorbic acid concentrations do not exceed 100 µM, matching the affinity and capacity of SLC23A1 isoform 1B [11], rendering an additional isoform obsolete. This would be consistent with the general theory that alternative splicing multiplies the encoding volume of any given genome of eukaryotes and the differential regulation of alternative splice variants is considered to play a substantial role in defining the phenotype of a given cell type, or cell state [23].
To conclude, this is the first report of an alternative splice variant since the initial description of the SLC23A1 transcript [11,12,24] or the characterization of the gene locus [13,14]. The novel alternative protein isoform A exhibits low ascorbic acid transport activity at a substrate concentration of 300 µM, however, its specific roles and possible synergism with the more functional SLC23A1 isoform 1B remains to be determined. Moreover, the human SLC23A1 locus extends about 1kb further 5’ than previously described [13,14].
ACKNOWLEDGEMENTS
This work was funded through Peter Eck’s Canada Research Chair in Nutrigenomics award and in part by the National Institute of Diabetes and Digestive and Kidney Diseases intramural program grant numbers Z1A DK053218 06 and Z1A DK054506 15.
![The novel SLC23A1 isoform 1A mediates ascorbic acid uptake in Xenopus laevis oocytes. [14C]ascorbic acid uptake into Xenopus laevis oocytes injected with cRNA encoding SLC23A1 isoform 1A [?] is elevated compared to sham injected oocytes [?], however, it is only about 7% compared to SLC23A1 isoform 1B [?], when incubated with 300 ?M [14C] ascorbic acid.](https://www.jscimedcentral.com/public/assets/images/uploads/image-1707219865-1.png)








































































































































