Molecular Characterization of Some Nigerian Date Palm Accessions Using RAPD Markers
- 1. Nigerian Institute for Oil Palm Research, Date palm Research Substation, Dutse, Nigeria
- 2. Department of Plant Biology, Federal University of Technology, Minna, Nigeria
- 3. Department of Biological, Geological an Environmental Sciences, Cleveland State University, Ohio, U.S.A
- 4. National Biotechnology Development Agency, Agricultural Biotechnology Department, Abuja, Nigeria
- 5. Department of Plant Science and Biotechnology, Nassarawa State University, Keffi, Nigeria
- 6. Department of Biological Sciences, Federal University Dutse, Nigeria
Abstract
Date palm (Phoenix dactylifera L.) belongs to Arecaceae family. It is a dioecious and long lived monocotyledonous fruit tree and one of the most important tree crops of Sahel, Sudan and Guinea Savannah ecologies in Nigeria. The genetic diversity of the crop in the experimental gene pool conserved in Dutse, Nigeria, is yet to be ascertained, most especially, using molecular markers. The leaves of each of the 21 accessions were collected from young (3 weeks-1 month old) date palm seedlings. A total of 125 amplified fragments were distinguished across the selected primers and the statistical analysis showed 106 polymorphic bands among the 21 genotypes with an average of 84.3 polymorphic bands per primer. The highest numbers of fragment bands were produced by the primer OPH-06 with 100% polymorphism while the lowest numbers of fragments were produced by the primer OPH-04 with 71% polymorphism. Cluster analysis by UPGMA showed 12 main clusters among the genotypes based on difference at the genetic level. The highest diversity (least similarity coefficient, 0.07) was found between accessions R5P24 and R16P31 in group IV, indicating low genetic relationship and high genetic diversity between the accessions. The least diversity (highest similarity coefficient, 4.04) was found between accessions ZARIYA and R4P29 in group III. The results of molecular analysis were adapted with genetic variation. RAPD technology therefore proves very effective for identifying genotypes, polymorphism and genetic distance of date palm. Therefore, making it very useful for future breeding programmes.
Keywords
• Date palm
• Accessions
• Diversity
• Molecular markers
• RAPD
CITATION
Yahaya SA, Daudu OAY, Mariam NI, Muhammad LM, Okoye CA, et al. (2023) Molecular Characterization of Some Nigerian Date Palm Ac cessions Using RAPD Markers. J Hum Nutr Food Sci 11(4): 1174.
INTRODUCTION
Date palm (Phoenix dactylifera L.) belongs to Arecaceae family. It is a dioecious and long lived monocotyledonous fruit tree in arid and semi-arid regions in the world with high tolerance to environmental stress. Great diversity exists in date fruit size, color, quality, quantity and harvesting time. Among them, only a few cultivars have immense value in maintaining and enhancing the quality of date palm cultivation, agro management and commercialization of this crop [1]. Slow growth, dioecy, slow offshoot-based clonal propagation system and impossibility of evaluation of genetic diversity on the basis of seedling morphological characteristics have severely restricted the improvement program of date palm cultivation.
Molecular markers have been used to evaluate genotypic and phenotypic variation in date palm in the last decade. A gene of interest can be linked from molecular markers thereby allows identification of commercial varieties and indirect selection of desired genotypes [2] DNA markers have yielded good results in some breeding programmes [3] and in studying crop genetic relationships [4]. Differences at the gene or DNA sequence level can be detected by various molecular marker systems (isozymes, RFLP, AFLP, RAPD, SSR, SRAP, CAP). With these markers, gene identification, genetic diversity determination, and chromosome and trait mapping can be done.
In order to determine polymorphism between plant accessions or species; molecular markers are used. Polymorphism detection is important for breeding of new plant cultivars. Molecular marker systems are more effective than morphological marker systems to define genetic relationship among watermelon cultivars. In early work, isozymes were used to determine genetic diversity and phylogenetic relatedness among watermelon cultivars.
MATERIALS AND METHODS
Molecular Analysis
The molecular work was carried out at Bioscience Center, International Institute for tropical Agriculture (IITA)
Sample Collection
The leaves of each of the 21 accessions were collected from young (3 weeks- 1 month old) date palm seedlings. The leave samples were dried using silica gel and taken to IITA, Ibadan for analysis.
DNA Extraction
The DNA extraction using a CTAB (Cetyltrimethyl ammonium bromide) based procedure using DNeasy plant mini kit (Qiagen, Germany), according to the manufacturer’s instructions.
DNA Amplification Protocol and Primer Selection
The PCR (Polymerase chain reaction) programme included an initial denaturation step at 94°C for 2 mins followed by 45 cycles with 54°C for 1 min for annealing 30 seconds, extension at 72 °C for 30 seconds and final extension at 72oC for 10 minutes was carried out.
A total of six primers were tested with the twenty-one accession of date palm (Phoenix dactylifera). The six informative primers were selected and used to evaluate the degree of polymorphism and genetic relationship between and within all accessions under study.
Electrophoresis
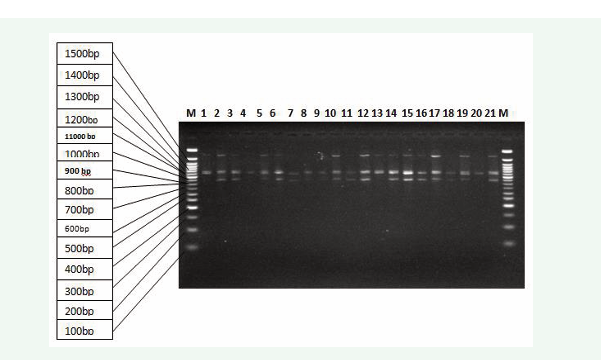
The amplified DNA fragments were separated on 1.5% agarose gel and stained with ethidium bromide. Base pair (bp) DNA ladder (Progma) was used as a Marker within molecular size from 100-1500 bp. Visualized under 300 nm UV light after staining with ethidium bromide.
Data Analysis
The data collected for morphological and yield parameters, proximate analysis, mineral composition, phytochemical screening in all the accessions were subjected to analysis of variance (ANOVA) using SPSS. Duncan Multiple Range Test (DMRT) was used to separate the means where significant difference was observed.
For molecular Analysis, Binary data was generated for each primer sets using 1 (presence of positive amplification at a particular band size) and 0 (absence of positive amplification at a particular band size) the generated binary data was the used to create a data matrix which was analyzed using the Power marker V2.35 software.
Cluster analysis, a method for displaying differences or similarities among cultivars, was employed for grouping together genotypes that showed dissimilarity in several traits. Clustering was carried out using the unweighted pair--group method of arithmetic average (UPGMA) method. All statistical analyses were performed using Statistical Analysis System software V6.07.
RESULTS AND DISCUSSION
The genetic distances among the twenty-one date palm accessions obtained using UPGMA are represented below. The obtained dendrogram separated the date palm accessions into 6 main Clusters, Based on the clustering, group I consist of four sub clusters representing 19% of the accessions which are R24P9, R4P12, R5P8 and R5P20, Group II also had four sub clusters representing 19% of the accessions which includes the accessions R13P9, R3P22, R2P4, R13P1, Group III , Group IV and Group V had two sub clusters each representing 10% of the total accessions, these accessions includes (ZARIYA, R4P29), (R5P24, R16P31) and (R13P5 and R7P1). Group VI had the highest number of sub clusters with a total number of seven accessions, these represents 33% of the total accessions. These accessions include R1P10, R6P20, R9P12, R15P6, R9P2, R14P21 and R1P18 respectively.
The jacquard similarity index for the calculation of distances among the date palm accessions revealed an interesting phenomenon. A distinct relationship and differences among the accessions was found. The highest diversity (least similarity coefficient, 0.07) was found between accessions R5P24 and R16P31 in group IV, indicating low genetic relationship and high genetic diversity between the accessions. The least diversity (highest similarity coefficient, 4.04) was found between accessions ZARIYA and R4P29 in group III.
Primer selection and DNA Amplification
The result indicates that the six (6) primers showed at least one (1) consistent polymorphic band. A total of 125 amplified fragments were distinguished across the selected primers and the statistical analysis showed 106 polymorphic bands among the 21 genotypes with an average of 84.3 polymorphic bands per primer.
The highest numbers of fragment bands were produced by the primer OPH-06 with 100% polymorphism while the lowest numbers of fragments were produced by the primer OPH-04with 71% polymorphism. Polymorphic Information Content (PIC) values ranged from 0.616 to 0.920 (Table 1). Moreover, the size of the amplified fragments varied with different primers, ranging from 100 to 1500 base pairs. The polymorphisms revealed by the six (6) primers indicate that they are good and reliable for genetic diversity assessment in date palm (Phoenix dactylifera) and there is a high degree of diversity in the accessions studied.
This study has reflected that genetic diversity is available among date palm genotypes from Northern Nigeria. The phylogenetic tree constructed revealed that date palm accessions examined provided evidence of divergence since they were grouped in clusters. This confirms with the findings of [5], on Phylogenetic relationships among date palm (Phoenix dactylifera L.) cultivars in Syria using RAPD and ISSR markers, similar findings were also reported by [6], On Molecular characterization of some Egyptian date palm germplasm using RAPD and ISSR markers.
The high level of polymorphism (84.5%) observed among the accessions is analogous to the 83% polymorphism noticed in a study of genetic diversity in some Iraqi date palms as reported by [7]. An example of genetic polymorphism across the 21 date palm accessions using primer OPH-06 and OPH-10 are shown in Table 2 respectively. The high polymorphism observed is an indication of prevalence of good diversity among the accessions studied in this present investigation.
Results also showed that some accessions from the same gene pools are grouped together, e.g. accessions R9P12, R6P20 and R1P10 as revealed in Figure 1,2,
Figure 1: Dendrogram showing the phylogenetic relationship among Twenty-one date palm accessions based on RAPD Marker
Figure 2: Gel Electrophoresis of the date palm accessions
This could be explained that the clustering together of accessions from the same gene pool are indications that genetic divergence followed geographical separation. Reported that date palm had a common evolutionary relationship but some accessions have adapted well to their local environments through gene rearrangement due to long periods of cultivation, making them to become landraces.
However, in group II, the UPGMA revealed slight intra varietal polymorphism among the accessions from the gene pools and did not group the accessions belonging to each of the gene pools to different groups, making their genetic polymorphism to be narrow. This reflects the close relationships among the accessions and suggests the occurrence of gene flow. Similar scenario on some date palm was observed in Saudi Arabia based on RAPD and ISSR markers [8], on some Tunisia date palm using RAPD Marker [9], this has also been justified by [10] on Molecular phylogeny of date palm cultivars from Saudi Arabia, and suggested that the ex-change of the varieties between the different plantation areas, clonal propagation of ecotypes and development of new recombinants by seedling selection and limited sexual reproduction may have been the main reason for less diversity among them.
Genetic diversity can be enlarged by combining desired traits from different local and wild populations of different geographical origins into the breeding lines [11]. However, it may be appropriate to analyze accessions that represent a wide range of more geographical origins in order to maximize genetic diversity. Nevertheless, the rich genetic diversity available among the date palm accessions from Northern Nigeria can be utilized for current and future breeding programs in order to select genetically distinct parents.
The use of RAPD for assessment of genetic diversity is preferred over the morphological and biochemical markers since these are completely devoid of the effects of environment and the stage of the experimental material, thus making them highly reliable [12]. Reported that it is simple to use for the evaluation of genetic diversity and phylogenetic studies in many plant species. This has been confirmed by several authors [13].
The present study also confirmed the suitability of RAPD as a reliable, simple, easy to handle and elegant tool in molecular diagnosis of different accessions available in the germplasm collection. The results from this study have revealed that RAPD markers can efficiently evaluate genetic variation in date palm germplasm.
CONCLUSION
The current study implies that molecular markers as RAPD are effective tools to discriminate various date palm genotypes. Molecular markers are absolutely necessary for modern breeding to achieve date palm genetic variation improvement considering the lengthy and dioecious nature of date palm. Even though the germplasm gene pool conserve in NIFORs date palm research substation has the largest diversity of date palm in Nigeria, further research should be done on other molecular markers for better overage on date palm trees that are not included in this present study. Until now, all the accessions of date palm are not registered properly. Furthermore, there will be genetic erosion due to expansion of improve varieties, thus proper registration should be one on local landraces.