Guanidino Compounds as Kidney Disease Toxins
- 1. Health Research Foundation, Research Institute for Production Development 4F, Japan
ABBREVIATIONS
CKD: Chronic Kidney Disease; KF: Kidney Failure; CTN: Creatinine; G: Guanidine; MG: Methylguanidine; GSA: Guanidinosuccinate; CSF: Cerebrospinal Fluid; GAT: Glycine Amidinotransferase; CK: Creatine Kinase; ARG: Arginine; PDB: Protein Data Bank; Adome: S-Adenosyl Methionine; ORN: Ornithine; Adohey: S-Adenosyl-L- Homocysteine; AMP: Adenosine Mono-Phosphate; ADP: Adenosine Di-Phosphate; ATP: Adenosine Tri-Phosphate
INTRODUCTION
Early detection of personal abnormal metabolism and treatment of weakened people are easy and economical ways to keep people healthy. The self-control of individual health is essentially necessary to keep them healthy. A typical example is diabetes in the USA. We can easily observe the very fat and pre-diabetic people in the USA. Such poor culture is spreading in the world. Kidney disease is a part of poor self-control results. The early- stage monitoring of personal metabolisms can prevent kidney disease. The biological evidence of chronic kidney disease (CKD) has been described in detail. Solutes in uremia were classified into three groups: high, middle, and lower (<500 Dalton), and the lowest group contains 68 compounds, including urea and creatinine, and uremic toxins and toxicity were studied. Many >500 Dalton compounds were found for further study [1]. The causal relationship between metabolites and CKD was unclear, and 48 metabolites were to be associated with various outcomes. N- acetylleucine was a potential marker of kidney tubular function [2], had a lower risk of CKD, and the glycine- alanine ratio had a higher risk of CKD [3]. An unnamed metabolite in urine and plasma was significantly associated with kidney failure (KF) [4], and the compound K extract from Ginseng Sprouts, C-glucosyltryprophan, a metabolite belonging to the phosphatidylcholine pathway, improved the prediction performance for KF [5]. Fumarate, allantoin, robotic acid, and nucleotide were associated with mortality in CKD [6]. The urea cycle, the arginine- nitric oxide pathway, the polyamine pathway, and short- chain acylcarnitine metabolites were altered in adults with CKD and impaired renal function [7]. The progression of CKD was closely associated with systemic inflammation and oxidative stress. Certain minerals, vitamins, and plant-derived metabolites exhibited beneficial effects [8]. Potential markers of glomerular filtration (pseudouridine), histamine metabolism (methylimidazolacetate), and azotemia (homo-citrulline) were in fully adjected models in African-American Study of a Kidney Disease and Hypertension and Atherosclerosis Risk in Communities [2].
Furthermore, abnormal microbiota composition and metabolites, in turn, participated in developing CKD. Altered gut microbiota and its metabolites might lead to significant changes in metabolic homeostasis and inflammation, ultimately contributing to the onset of sarcopenia [9], and mitochondrial fatty acid oxidation- regulated monocytic type 1 interferon signaling via histone acetylation [10]. Impaired kidney function and CKD leading to kidney failure and end-stage renal disease risk was a serious medical condition associated with increased metabolic disturbances, including hypertension, dyslipidemia, and anorexia-cachexia syndrome, which were linked to poor outcomes. Specific hormonal, inflammatory, and nutritional-metabolic factors might play key roles in CKD development and pathogenesis [11], and specific plasma metabolites causally associated with CKD and renal functions provide potential targets for the intervention [12]. CKD was a debilitating pathology with various causal factors, culminating in end-stage renal disease requiring dialysis or kidney transplantation. CKD leads to microglial potassium efflux and inflammasome activation in the brain [13].
On the other hand, several guanidino compounds have been found in the serum of nephritic patients [14], and may play an essential role in the etiology of uremic encephalopathy. Four guanidino compounds appeared to be highly increased in serum, cerebrospinal fluid, and brain of uremic patients. Creatinine (CTN), guanidine (G), guanidinosuccinate (GSA), methylguanidine (MG), and some guanidino compounds have been suspected to be uremictoxins [15]. MGwas shown toberelatedtotheuremic polyneuropathy found in uremia [16]. The excitatory effects of uremic guanidino compounds on the central nervous system may be explained by the activation of N- methyl- D-aspartate receptors by GSA, concomitant inhibition of GABA [9], receptors by uremic guanidino compounds, and other depolarizing effects [17]. GSA, MG, G, and CTN were suggested to be the cause of chronic and generalized seizures after systemic and intracerebroventricular administration in mice [18,19]. GSA was related to uremic bleeding diathesis and has been shown to inhibit excitatory neurotransmission in rat hippocampal brain slices, an effect that hypothetically could contribute to uremic encephalopathy [20]. The kinetics of GSA in the circulating blood using a rat model of cisplatin-induced renal failure and guanidino succinate transport between the circulating blood and GSA. The renal elimination of GSA is attenuated in cisplatin-treated rats. The greater elimination clearance of GSA from CSF may contribute to the maintenance of a low GSA concentration in the serum and CSF [21]. The effects of clinical dialysis have been studied by measuring the guanidino compounds by liquid chromatography. The serum guanidino compound levels found in uremic children were compatible with those in adults. Single hemodialysis lowered the level of most serum guanidino compounds temporarily, and the level remained more stable in continuous-cycle peritoneal dialysis-treated patients [22]. The measurement of L-arginine, which is a physiological precursor of nitric oxide, was used in studies of the disposition of exogenous doses of L-arginine [23]. A mechanistic explanation for the differences in the extent of decreases in the levels of different guanidino compounds was unavailable. However, no correlation was reported between the molecular weight or the acidity of the guanidino compounds and their percentage decrease. The guanidino compounds were distributed throughout the tissue, and their binding to protein and other factors contributed to the differences in their dialysability [24].
HPLC analysis of guanidino compounds found a large quantity of arginine and creatine in kidney disease patients’ sera. If people do not take ARG and CT as supplements, the enzyme activity of glycine amidinotransferase (GAT) and creatine kinase (CK), are weakened by poor production or third-party inhibition [25]. The possible treatment is to dose RNA of these enzymes as a molecular level reproduction or activate these enzymes by dose suitable compounds; therefore, we must develop a fast diagnosis of these enzymes. The approach is either directly analyzing the enzyme activity or studying the precursor and metabolite. First, we have to make clear the reaction mechanism to focus the target compounds for the development of fast analysis.
EXPERIMENTAL
These computational chemical analyses were performed using Molecular Mechanics to optimize the structures and MOPAC PM5 programs to calculate the atomic partial charge of SciGress supplied by Fujitsu Japan with a desk-top computer equipt a 3.2 GHz cpu. The enzyme reaction mechanism was analyzed using the enzyme stereo structures from RSCB PDB after the downloaded stereo structures were corrected, especially for the substitutes.
RESULTS
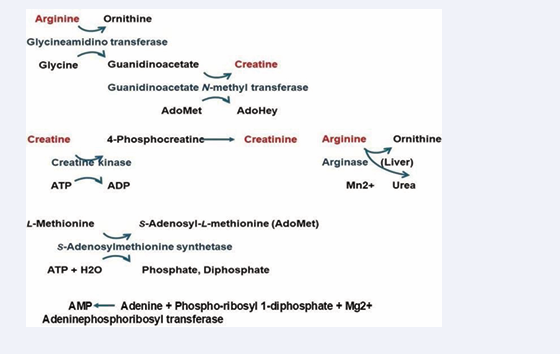
Analysis of enzyme reaction mechanisms related to guanidino compounds was performed in silico. The metabolism of arginine to creatine is the three-step enzyme reaction. The first reaction is from arginine to ornithine by glycineamidinotransferase, the second step reaction is from guanidinoacetate to creatine by guanidinoacetate N-methyltransferase, and the third reaction is from creatine to 4-phosphocreatine by creatine kinase. The analysis processes and results are explained in the individual enzymes. The replacement of amino acids and the replacement and insertion of cofactors of downloaded PDB proteins were necessary to explain these human enzyme reactions. The primary metabolism of a guanidino compound from arginine to creatine was quantitatively analyzed and visualized. The target enzymes were glycineamidino transferase, guanidinoacetate N-methyl transferase, and creatinine kinase. Figure 1 shows the metabolic pathway.
Figure 1: Metabolism scheme from arginine to creatinine
Glycineamidino transferase
The metabolism (reaction) mechanism from glycine to guanidinoacetate was described as the contribution of the enzyme serine355 (S355), where the serine contacted with the arginine guanidino group and the guanidino group was transferred to glycine [26]. However, S355 hydroxy group contacts with the substrate arginine carboxy group (4JDW) [27], and other substrates r-aminobutyric acid (6JDW), norvaline (1JDX) carboxy group contacts with the serine hydroxy group and their amino groups are free from contact with the serine. Another proposal about the reaction mechanisms is glycine amino group contacts with the substrate arginine guanidino group, and the guanidino group transfers to glycine [27,28]. The reaction mechanism was examined in silico using downloaded GAT structures.
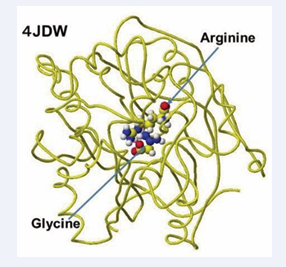
The several stereo structures of human GAT were downloaded from RSCB PDB, and 4JDW was chosen due to the substrate arginine. The stereo structure was corrected and optimized using a CAChe# MM2 program. The residual amino acids within 3Å were extracted for their further analysis. A glycine molecule was installed near the arginine guanidino group and optimized to study the feasibility of electron transfer from arginine guanidino to the glycine. The atomic distance and atomic partial charges of target atoms support the possibility of their contact. The downloaded, corrected, and optimized structure of GAT (4JDW) is shown in Figure 2.
Figure 2: Downloaded and optimized stereo structure
The location of arginine and glycine are indicated as their molecular form, and the protein is exhibited as line form.
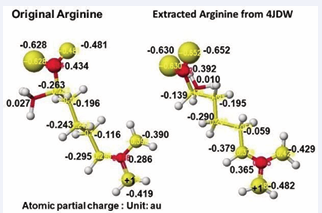
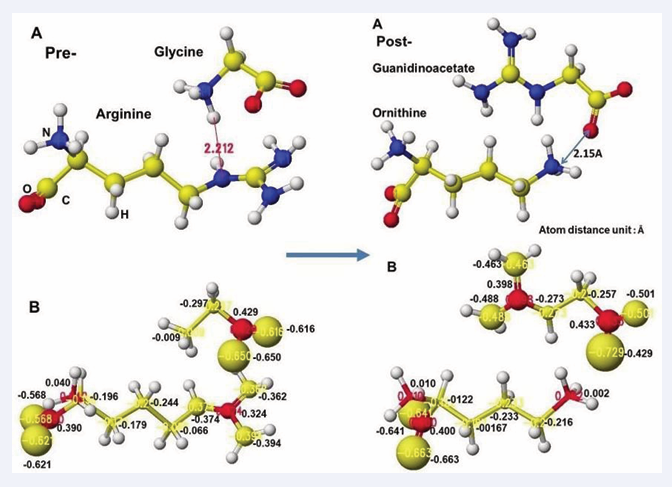
According to the GAT enzyme reaction mechanism report, the protein’s serine removes arginine’s guanidino group, transfers to a glycine amino group, and produces guanidinoacetate. However, the stereo structure indicates that the carboxy and amino groups are located near the residual arginine guanidyl and serine hydroxy groups, the substrate arginine guanidino group is far from the molecular interaction site, and no serine exists near the substrate arginine guanidino group. The stereo structure indicates that rotation of the substrate arginine is difficult in vivo reaction. Therefore, the electron localization of the substrate arginine before and after being trapped in the enzyme reaction center was analyzed. Figure 3 shows the extracted substrate arginine and original arginine.
Figure 3: Atomic partial charge of arginine heavy atoms of Arginine: Glycineamidinotransferase (GAT) of free form (original) and optimized inside the enzyme including arginine and glycine
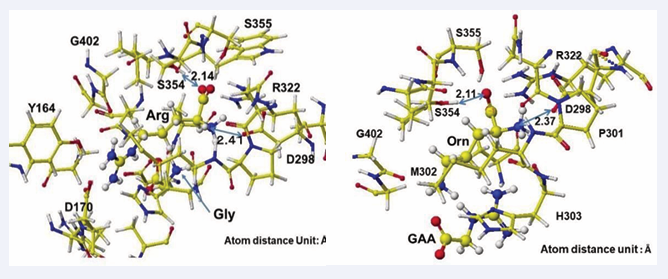
Figure 3 shows the extracted conformation of substrate arginine by surrounding residual amino acids from 3Å distance from the substrate arginine. The feasibility of enzyme reactions is the same as that of chemical reactions. The localized electron of atoms near the reaction center indicates the reactivity; therefore, arginine’s atomic partial charge (apc) before and after was calculated using a MOPAC PM5 program. The arginine carboxy group contacts the residual S355 and R322, and the amino group contacts the residual D298. The substrate arginine is tightly held by the residual amino acids S355, R322, and D295 and cannot be rotated inside the reaction chamber, and the guanidino group can contact glycine. The electron localization explains the binding tightness. The atomic partial charge values of target atoms shown in Figure 4 indicate the binding center.
Figure 4: Extracted stereo structure of arginine and substitutes with residual amino acids within 3A from the arginine (Arg) or ornithine (ORN). The key atom distances are indicated in the Figures.
The complex structures of arginine and glycine complex and ornithine guanidinoacetate are shown in Figure 5.
Figure 5: Extracted conformations of pre- and post-reactions with heavy atom atomic partial charge values.
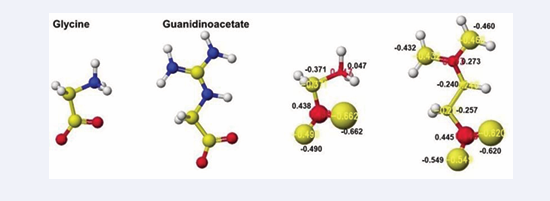
Initially, glycine tightly contact with the arginine guanidino group (Figure 5A Pre), and the arginine guanidino group is transferred to the glycine amino group; then, the arginine is converted to ornithine, and the glycine becomes guanidinoacetate. The electron localization is shown in Figure 5B. The atomic partial charge change indicates their contact center and the feasibility of the reaction. Figure 6 exhibits the original structure of glycine and guanidinoacetate with the atomic partial charge of heavy atoms to make clear the reaction center.
Figure 6: Optimized structure and heavy atom atomic partial charge values of glycine and guanidinoacetate
The apc values of the substrate arginine carboxy oxygen are -0.628 and -0.481, and those held by the enzyme are -0.652 and -0.630 au; the strong negative values are recovered a little by forming the complex with glycine. The apc value of guanidino carbon is changed from 0.286 to 0.365 au by the folding and a little recovered by the complex formation to 0.324 au.
The reactivity is also understood from the apc value change of glycine and guanidinoacetate. The apc values of glycine carboxy oxygen atoms change from -0.490 and-0.662 to -0.616 and -0.650 au. Those guanidinoacetate carboxy oxygen atoms change from -0.620 and -0.549 au to -0.724 and -0.501 au. Such conformation analysis and apc value change make clear the reaction center and the enzyme selectivity.
Guanidinoacetate N-methyltransferase
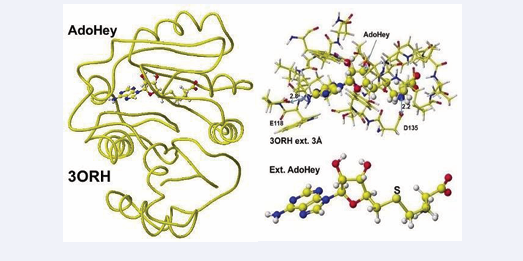
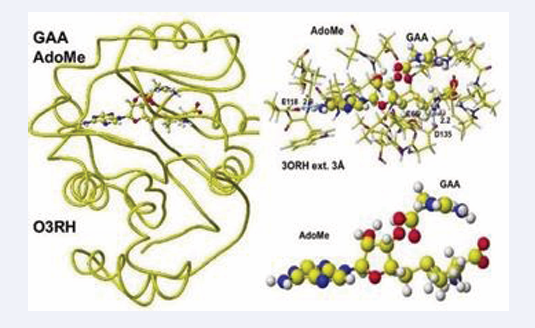
The enzyme activity is damaged by both inborn error and disease. Early oral dosing of creatine can normalize inborn error patients [29], and the weakened GAT activity is determined by analysis of GAA in urine and sera; however, one spot analysis is affected by personal lifestyle and requires balanced analysis of metabolites. The following enzyme after GAT is guanidinoacetate N-methyltransferase, which metabolizes guanidinoacetate to creatine. The coenzyme is AdoMe, whose S-methyl group is transferred to the guanidinoacetate guanidyl group. A human guanidino N-methyltransferase is downloaded from the PDB (3ORN), including S-adenosyl-L- homocysteine (AdoHey). The optimized structure, extracted structure of AdoHey with residual amino acids within 3Å, and extracted AdoHey structures are shown in Figure 7.
Figure 7: Downloaded and optimized the structure of Guanidinoacetic acid N-methyltransferase, extracted and S-adenyl-L-homocysteine.
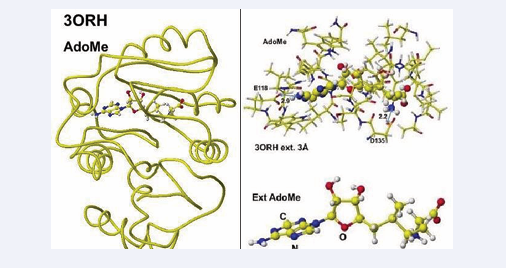
The AdoHey carboxy group binds with the Aspartic acid (D135) carboxy group via ion-ion interaction; the atomic distance between amino nitrogen carboxy oxygen is 2.2Å. AdoHey adenosine amino group binds with glutamic acid (E118) carboxy group; the atomic distance between the nitrogen and oxygen is 2.9Å. These bonds keep the AdoMe and AdoHey structure straight. The atomic partial value of the key atom sulfur changed from 0.869 to 0.914 au, and that of methyl group carbon did from -0.447 to -0.572 au by electron localization, First, S-adenosyl-L-homocysteine is converted to S-adenosyl methionine, a guanidinoacetate is inserted, and the chemical structures are corrected. The optimized complex structure is shown in Figure 8,
Figure 8: Optimized structure of Guanidinoacetic acid N- methyltransferase with modified S-adenosylmethionine and the extracted conformation and S-adenosylmethionine.
including residual amino acids within 3Å from AdoMe and extracted AdoMe. The holding of AdoMe inside the enzyme is the same as that of AdoHey. Insert GAA; then, the complex structure is optimized. The GAA carboxy group contacts the AdoMe sulfur. The construction of the transition state from GAA to creatine is difficult inside an enzyme. Therefore, the docking of creatine with AdoHey is performed by the insertion and the whole structure optimization. The result is shown in Figure 9, including an extracted conformation with residual amino acids with 3Å from AdoHey.
Figure 9: Optimized structure of Guanidinoacetic acid N-methyltransferase, S-adenosylmethionine, and guanidinoacetate, extracted conformation, and S-adenosylmethionine guanidinoacetate complex
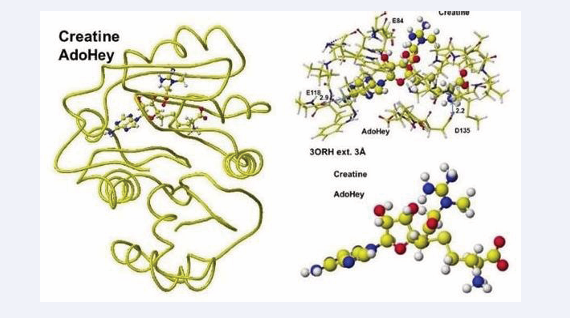
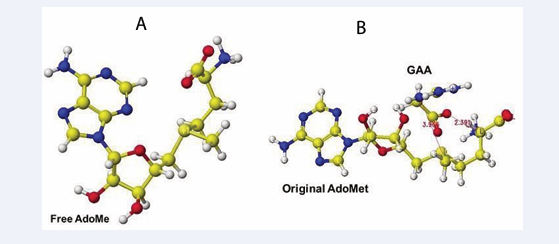
The extracted and modified AdoMe is bent outside the enzyme after the optimization, as shown in Figure 10.
Figure 10: Optimized structure of Guanidinoacetic acid N-methyltransferase with S-adenosyl-L-homocysteine and creatine complex, the extracted conformation, and S-adenosyl-L-homocysteine and creatine complex.
Therefore, the docking process is not automated, and guanidinoacetate and creatine are carefully inserted and optimized complex structures. Guanidinoacetate and creatine carboxy group binds with S-adenosylmethionine and S-adenosyl-L-homocysteine amino group in vitro (Figure 11),
Figure 11: Optimized structure of S-adenosylmethionine (A), Complex of extracted S-adenosylmethionine and guanidinoacetic acid (B).
but the amino group already binds with the residual amino acid carboxy group in vivo; therefore, the guanidinoacetate and creatine carboxy group locates rear sulfur atom of these coenzymes.
Creatine kinase
One of the bio-markers of diagnosis in kidney disease is creatine kinase, which metabolizes creatine to creatine phosphate, which is automatically metabolized to creatinine [30]. Therefore, creatine kinase activity can be determined by the concentration ratio of creatine and creatinine. Creatine is taken as a supplement, and the concentration in sera is affected by personal lifestyle; therefore, measuring the balanced amount of creatine and creatinine is necessary to determine the individual health condition.
Several stereo-structures of human creatine kinase are downloaded from RSCD PDB and corrected the structures, then optimized using an MM2 program. Human brain creatine kinase PDB 3DRB [31], includes ADP, Mg, and nitrate; however, human brain creatine kinase 3DRE contains (-)-7-[(6-chloro-2-naphtalemyl)-1’- (pyridinyl)- spiro[5H-oxazolo[3,2-a]pyrazine-2(3H), 4’-priperidin]-5- one instead ADP even though the report indicates ADP as a cofactor. It seems that the stereo structure of 3DRE [26], is deformed; therefore, human creatine kinase PDB 7TUN [32], is used as the standard structure after comparison with other human creatine kinases. An initial complex conformation with cofactors is critical for optimizing the complex structure; therefore, the PDB 4RF9 [33], structure is used as a reference structure.
Human sarcomeric mitochondrial creatine kinase 4ZPM includes ADP and uranium and has 72.4% similarity to human brain creatine kinase 7RUN and T. Californica Creatine Kinase PDB 1VRP containing ADP, guanidinoacetic acid, magnesium, and nitrate [34], has 89.3% similarity to 7TUM, and their secondary structures are identical. Therefore, 7RUM is used as the standard for human brain creatine kinase, and 1VRP is used for further analysis to visualize the human creatine kinase reaction mechanism. Therefore, the location of ATP and ADP is carefully managed by comparison of several crystal structures. First, 1VRP ADP is converted to ATP following the stereo structure of Anthoplasma PDB 4RF9 [33], and the residual amino acids of 1VRP are replaced with the residual amino acids of 7TUM. The mutation process is the same as that created by a human D-amino acid oxidase from east D-amino acid oxidase [35].
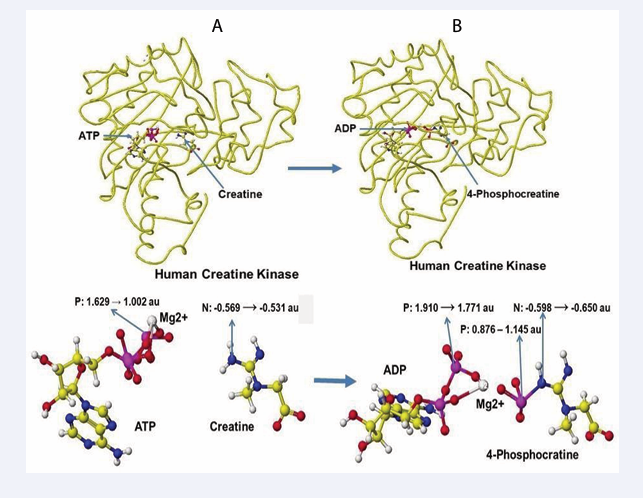
The optimized stereo structure of human brain creatine kinase is shown in Figure 12A. An ATP, a creatine molecule, and an ionized magnesium (2+) are included in the stereo structure. Some magnesium binds the hydroxy groups in reference structures, but the magnesium binds only phosphate oxygen in these structures. Magnesium has a crucial role in ATP activity [30,31], binds with three phosphate oxygens, and moves to bind with two remaining phosphate oxygens after releasing one phosphate to creatine. After releasing one phosphate to convert creatine to creatine phosphate, ATP converts to ADP. An optimized stereo structure of human creatine kinase with ADP and creatinine phosphate is shown in Figure 12B.
Figure 12: Stereo structures of human creatine kinase before and after the enzyme reaction
The atomic partial charge of ATP γ-phosphorus is changed from 1.529 to 1.002 au by the existence of creatine, and the atomic partial charge of creatine nitrogen increased from -0.569 to -0.531 au. The induction of the electron contributes to the molecular interaction. The phosphorus transport of ATP to creatine produces 4-phosphocreatine can also explained by the change of atomic partial charge values of key atoms. The β-phosphorus atomic partial value was reduced from 1.910 to 1.771 au, and that of the previous γ-phosphorus of ATP is changed from 0.876 to 1.145, and that of nitrogen of 4-phosphocreatine is reduced from -0.598 to -0.650 au. The optimization of chemical structure using the MM2 program to calculate the molecular interaction energy and the MOPAC PM5 to calculate the electron localization makes clear the enzyme reaction mechanisms.
DISCUSSION
In the process of glycineamidino transferase reaction, the enzyme converts from arginine to ornithine and from glycine to guanidinoacetate; therefore, glycine is one cofactor. Dosing glycine is one treatment method and another possible dosing RNA of glycineamidino transferase to increase the amount of GAT. If the activity of amino acid oxidase is too high, we shall dose the inhibitor to keep glycine content constant. The analysis of glycine is also one target of diagnosis. Guanidinoacetate N-methyl transferase, converting from guanidinoacetate to creatine, requires a cofactor AdoMet, which will be a drug candidate and also a supplement. The dosing of glycine in the kidney and manganese in the liver accelerates arginine metabolism. AdoMet can accelerate guanidinoacetate metabolism to creatine. Glycine, Magnesium, and AdoMet are available as supplements; therefore, we have to measure their amount not to exceed. AdoMet is synthesized from L-methionine by S-adenosylmethionine synthetase. The cofactor is ATP.
Creatine is metabolized to creatinine by creatine kinase, and ATP is also a cofactor. Taking ATP and ADP is not recommended due to the instability of both ATP and the precursor ADP. The precursor of ADP is AMP, which is synthesized from adenine and phosphor ribosyl 1-diphosphate using adenosine phospho-ribosyl transferase with magnesium ion, which is an essential element in this synthesis process, and further modification may provide a friendly medicine.
The enzyme reaction activating compounds and the increasing enzyme mass by RNA are the medicine candidates. The possible treatment of the disease is to dose RNA of these enzymes as a molecular level reproduction or dose suitable cofactors to activate these enzymes; therefore, we have to develop a fast personal diagnosis of these enzymes. The approach is either the direct analysis of the enzyme activity or the analysis of the precursor and metabolite. The analysis of glycine is also one target of a diagnosis test. If an inventor damages the enzyme activity, we should kill viruses and bacteria using anti- bacteria (antibiotic) and virus (antiviral) drugs. First, we need a precise diagnosis that indicates individual enzyme activities.
Monitoring these enzyme activities should help to prevent kidney disease. However, the easy availability of arginine and creatine as supplements causes the measuring actual amounts (concentration) of these compounds to make it difficult for individual diagnosis determination. Therefore, the amount of these compounds and their metabolites (ornithine and 4-phosphocreatine) should be analyzed, or analyzing cofactor glycine, guanidino-acetic acid, and creatinine is required. Taking L-methionine is one idea, but the enzyme S-adenosylmethionine synthetase requires ATP as the coenzyme, and creatine kinase also requires ATP to accelerate the activity. Therefore, the self- control of arginine and creatine metabolism is a very risky approach to controlling our health. We should avoid self- control and consult a medical doctor before taking these supplements. Chronic kidney disease is also crucial for the treatment; however, resistance exercise can improve patient health. Furthermore, in the critical subject of our aging society, the early identifying biomarkers of growing senile people and provide medicines to help MIBYO people, pre-dementia patients.
CONCLUSION
Visualizing enzyme reaction processes helps understand the enzyme reaction mechanisms, develop the diagnosis methods, and design medicines. However, personal lifestyle influences clinical analysis data such as arginine and creatinine those spot analyses do not provide individual health conditions. The biomarkers to determine kidney disease are arginine transferase and creatine kinase, and the measure of these enzyme activities can be determined by the relative values of arginine and guanidinoacetate and that of creatine and creatinine. Glycineamidino transferase converts (metabolizes) arginine to ornithine and glycine to guanidinoacetate, which is not a diagnosis target based on the total guanidino compounds analysis. The medical treatment candidates are the dosing enzyme reaction activating compounds and RNA to increase enzyme mass. ATP and ADP are unstable compounds; therefore, their precursors may be medicine candidates, and their further modification may provide friendly medicines. If an inventor damages the enzyme activity, we must kill viruses and bacteria using anti- bacteria. First, a precise diagnosis is required to determine individual enzyme activities.
REFERENCES
- Vanholder R, De Amet RI, Gloriuz G, Agile A, Baurmeister SU, Brunet P, Clark W, et al. Review on uremic toxins: Classification, concentration, and interindividual variability. Kidney Int. 2003; 63: 1934-1943.
- Wen D, Zheng Z, Surapaneni A, Yu B, Zhou L, Zhou W, et al. Metabolite profiling of CKD progression in the chronic renal insufficiency cohort study, JCI Insight. 2022; 7: e161696.
- Hou Y, Xiao X, Zhu Y, Li Y, Liu Q, Wang Z. Blood metabolites and chronic kidney disease: a Mendelian randomization study. BMC Med Genomics. 2024; 17: 147.
- Steinbrenner I, Schultheiss UT, Bachle H, Cheng Y, Schmid BM, Yeo WJ, et al. Associations of Urine and Plasma Metabolites With Kidney Failure and Death in a Chronic Kidney Disease Cohort. Am J Kidney Dis. 2024; 84: 469-481.
- Steinbrenner I, Schultheiss UT, Kotsis F, Schlosser P, Stockmann H, Mohney RP, et al. Urine Metabolite Levels, Adverse Kidney Outcomes, and Mortality in CKD Patients: A Metabolome-wide Association Study. Am J Kidney Dis. 2021; 78: 669-677.
- Hu JR, Coresh J, Inker LA, Levey AS, Zheng Z, Rebholz CM, et al. Serum metabolites are associated with all-cause mortality in chronic kidney disease. Kidney Int. 2018; 94: 381-389.
- Yamaguchi Y, Zampino M, Moadde R, Chen TK, Tian Q, Ferrucci L, et al. Plasma metabolites associated with chronic kidney disease and renal function in adults from the Baltimore Longitudinal Study of Aging. Metabolomics. 2021; 17: 1-19.
- Rapa SF, Di Iorio BR, Campiglia P, Heidland A, Marzocco S. Inflammation and Oxidative Stress in Chronic Kidney Disease- Potential Therapeutic Role of Minerals, Vitamins and Plant-Derived Metabolites. Int J Mol Sci. 2020; 21: 26.
- Zheng G, Cao J, Wang XH, He W, Wang B. The gut microbiome, chronic kidney disease, and sarcopenia. Cell Commun Signal. 2024; 22: 558.
- Wu JJ, Singh K, Shing V, Gupta A, Arenbergm BC, Huffstutler RD, et al. Mitochondrial fatty acid oxidation regulates monocytic type I interferon signaling via histone acetylation. Sci Adv. 2025; 11: 1-13.
- Slee AD. Exploring metabolic dysfunction in chronic kidney disease. Nutr Metab. 2012; 9: 36.
- Zhao X, Gao J, Kou K, Wang X, Gao X, Wang Y, et al. Causal effects of plasma metabolites on chronic kidney diseases and renal function: a bidirectional Mendelian randomization study. Front Endocrinol. 2024: 15:
- Zimmermann S, Mathew A, Bondareva O, Elwakiel A, Waldmann K, Jiang S, et al. Chronic kidney disease leads to microglial potassium efflux and inflammasome activation in the brain. Kidney Int. 2024; 106: 1101-1116.
- Takahashi Y, Kitasato Igaku, Hana T, Inamoto Y, Inamoto S. Analytical method to identify mal-function metabolites related to kidney diseases; Chromatography of guanidino compounds. J Chromatogr B. 2000; 747; 123-138.
- Bazilinsky N, Shaykh M, Dunea G, Mamdani B, Patel A, Czapek E, et al. Inhibition of Platelet Function by Uremic Middle Molecules. Nephron. 1985; 40: 423-428.
- Yokizaki T, Oura H, Ienaga K, Nakamura K. Effects of creatinine, creator and methylguanidine on renal function. J Nephrol. 1992; 34: 973-977.
- De Deyn PP, D’Hooge R, Van Bogaere PP, Marescau B. Endogeneous guanidino compounds as uremic neurotoxins. Kidney Int. 2001; 59: S77-S83.
- De Deyn PP, Macdonald RL. Guanidino compounds that are increased in cerebrospinal fluid and brain of uremic patients inhibit GABA and glycine responses on mouse neurons in cell culture. Ann Neurol. 1990; 28: 327.
- D’Hooge R, Pei YQ, Marescau B, De Deyn PP. Convulsive action and toxicity of uremic guanidino compounds: behavioral assessment and relation to brain concentration in adult mice. J Neurol Sci. 1992; 112: 96-105.
- D’Hooge R, Manil J, Colin E, De Deyn PP. Guanidinosuccinic acid inhibits excitatory synaptic transmission in Cal region of rat hippocampal slices. Ann Neurol. 1991; 30: 622-623.
- Kasai Y, Akanuma S, Kubo Y, Tachikawa M, Hosoya K. Pharmacokinetics of guanidinosuccinic acid in rat blood and cerebrospinl fluid. Drug Metab Pharmacokint. 2014; 29: 97-100.
- De Deyn PP, Marescau P, Swartz RD, Hogaerth R, Possemiers I, Lowenthal A. Serum guanidino compound levels and clearances in uremic patients treated with continuous ambulatory peritoneal dialysis. Nephron. 1990; 54: 307-312.
- Gopalakrishnan V, Burton PJ, Blaschke TF. High-performance liquid chromatographic assay for the quantitation of L-arginine in human plasma. Anal Chem. 1996; 68: 3520-3523.
- De Deyn PP, Robitaille B, Vanasse M, Qureshi IA, Marescau B. Serum guanidino compound levels in uremic pediatric patients treated with hemodialysis or continuous cycle peritoneal dialysis. Correlations between nerve conduction velocities and altered guanidino compound concentrations. Nephron. 1995; 69: 411-417.
- Hanai T, Inamoto Y, Inamoto S. Analytical method to identify mal- function metabolites related to kidney diseases; Chromatography of guanidino compounds. J Chromatogr B. 2000; 747: 123-138.
- Hanff E, Hafner P, Bollenbach A, Tsikas D. Effects of single and combined metformin and L-citrulline supplementation on L-arginine- related pathways in Becker muscular dystrophy patients: possible biochemical and clinical implications. Amino acids. 2018; 50: 6.
- Fritsche E, Humm A, Huber R. The ligand-induced structural changes of human L-arginine: glycine amidinotransferase. A mutational and crystallographic study. J Bio Chem. 1999; 274: 3026-3032.
- Tsikas D, Redfors B. Pilot study on acute effects of pharmacological intraperitoneal L-homoarginine on homeostasis of lysine and other amino acids in a rat model of isoprenaline-induced Taokotsubo cardiomyopathy. Int J Mol Sci. 2022; 23: 4734.
- Stockler-Ipsiroglu S, Apatean D, Battini R, DeBtpsse S, Dessofy K, Edvardson S, et al. Guanidinoacetatemethyltransferase (GAMT) deficiency: outcomes in 48 individuals and recommendations for diagnosis, treatment and monitoring. Mol Genet Metab. 2015; 116: 252-259.
- Hanai T. 2024-0529 Visualization XXIII Biomarker for Kidney Disease 1, Glycine amidinotransferase; 2024-0420 Visualization XVIII Possible RNA medicine candidates for kidney disease patients; 2024- 0326 Visualization XVI Quick search for advanced medicines; 2024- 0320 Visualization XV Advanced medicine, Small or Large molecules?
- Wang Z, Qiao Z, Ye S, Zhang R. Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains. Acta Crystallogr D Biol Crystallogr. 2015; 71: 779- 789.
- Lahiri SD, Wang PF, Babbitt PC, McLeish MJ, Kenyon GI, Allen KN. The A Structure of Torpedo Californica Creatine Kinase Complexed with the ADP-Mg (2+)-NO3(-)-Creatine Transition-State Analogue Complex. Biochemistry. 2002; 41: 13861-13867.
- Hanai T (ed) Quantitative in silico Analytical Chemistry, Quantitative Analysis of MolecularI from Chromatography Retention to Enzyme Selectivity, Chapter 9, Physical Biochemistry of Enzyme Reactions. 2024; 238-273.
Abstract
The enzyme reaction mechanisms related to guanidino compounds that are uremic toxins were analyzed using a computational chemical method (in silico analysis). The target enzymes for analyzing guanidino compound metabolism were Glycineamidino transferase, Guanidinoacetate N-methyltransferase, and Creatine kinase. Visualizing these enzyme mechanisms clarifies the reaction processes target clinical tests, drug candidates, and supplements. Dosing cofactors like glycine, AdoMet, and ATP should accelerate the metabolism reactions; however, the medicine candidate is limited due to the complex handling. The details of enzyme reactions and the reaction mechanisms are visualized for clear explanations.
KEYWORDS
- Kidney disease
- Guanidino compounds
- Uremic toxin
- In silico analysis
- Enzyme reaction
CITATION
Hanai T (2025) Guanidino Compounds as Kidney Disease Toxins. J Pharmacol Clin Toxicol 13(1):1187.